-
Supplementary File 1: Examples illustrating the
utilities of ToppGene Suite
a. Using ToppFun for gene list enrichment analysis
b. Using ToppGene for disease gene prioritization based on functional similarity to training set genes
c. Using ToppNet for disease gene prioritization based on topological features in PPIN
-
Supplementary File 2: List of diseases,
training and test set genes used to demonstrate the utility
of ToppGene and ToppNet
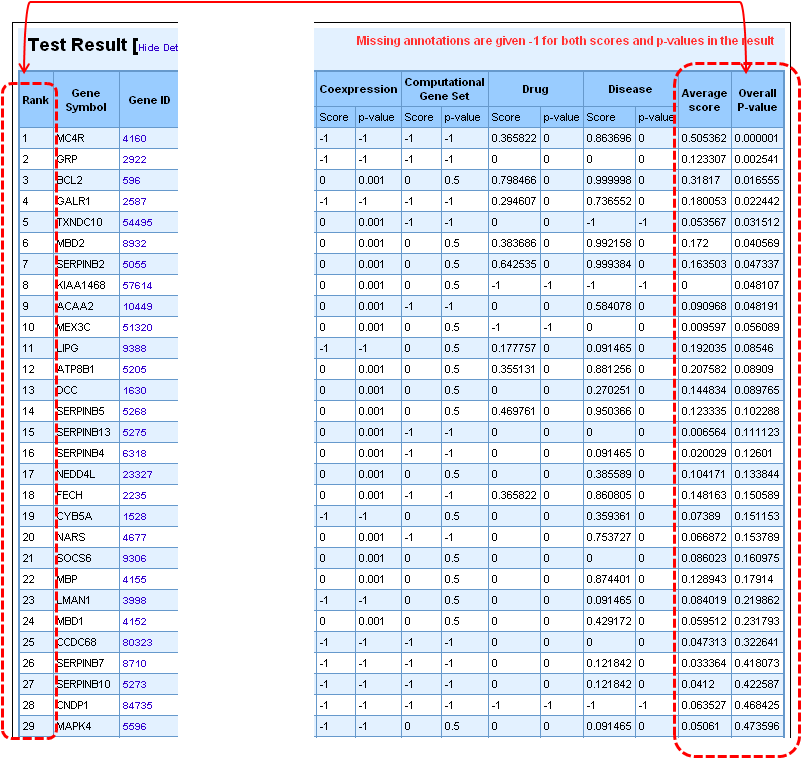
- Results of the 20 candidate gene prioritizations for the below listed five genetic diseases using ToppGene and ToppNet
- Bipolar Disorder
- Cardiomyopathy
- Celiac Disease
- Crohn's Disease
- Obesity
Supplementary File 1: Examples illustrating the utilities of ToppGene Suite
a. Using ToppFun for gene list enrichment analysis
Query: To conduct gene list enrichment analysis on obesity-associated genes.
1. From the homepage click on the first link ("ToppFun: Transcriptome, ontology, phenotype, proteome, and pharmacome annotations based gene list functional enrichment analysis")
2. On the "Gene list enrichment analysis" enter either human gene symbols or human NCBI Entrez Gene IDs in the box against "Training Gene Set" and click on "Submit Query". ToppGene supports RefSeq and UniProt IDs too. For the list of obesity genes, see the Supplementary File 2.
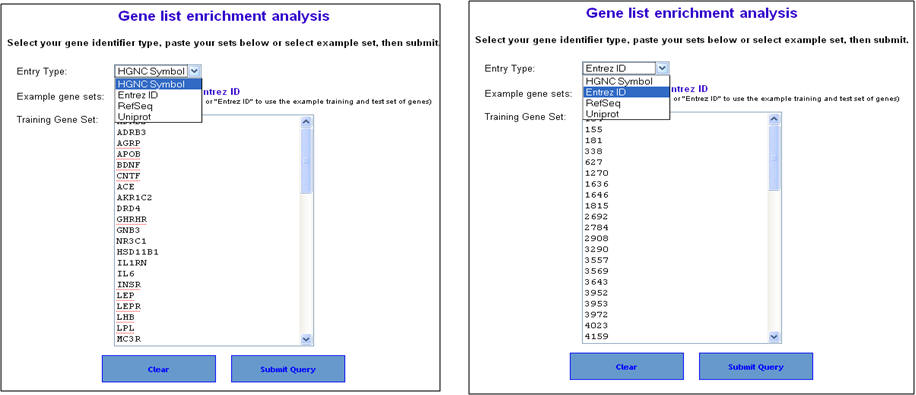
3. You can select the appropriate statistical parameters under the "Enrichment analysis parameter" and then hit "Submit" button. The default parameters are "Bonferroni" for multiple correction method and 0.05 for significance cut-off level. 4. If your gene list contains non-approved symbols or duplicates at this stage, they will be listed under "Genes Not found". You can then resolve this by selecting the appropriate symbols from a list of suggestions (see 4 below).
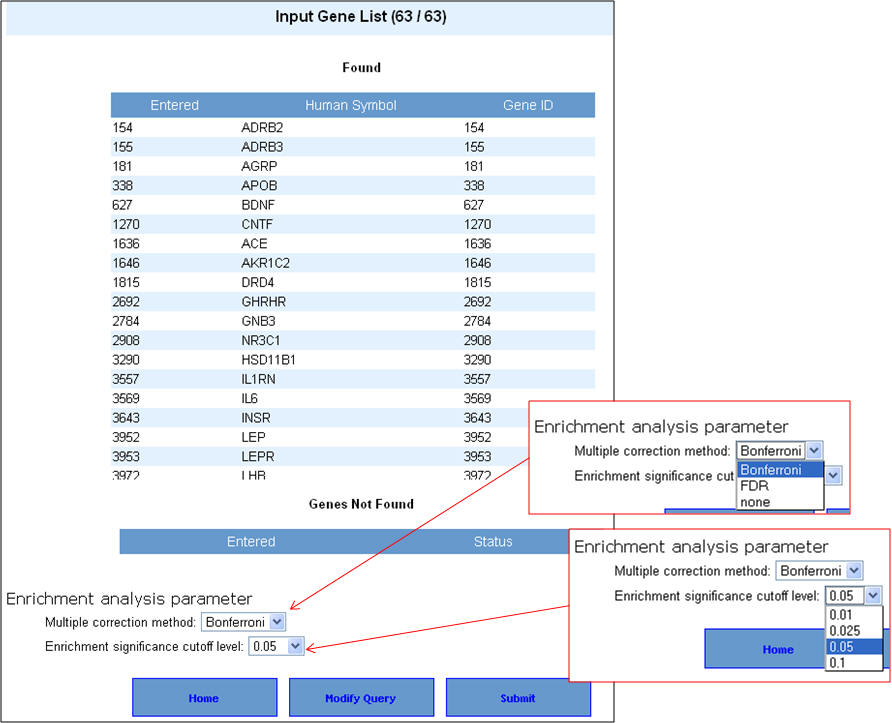
4. If your input gene list contains synonyms or alternate symbols other than approved human gene symbols, you will be prompted to select from a list of alternatives. Once you select the approved symbols, click on the "update" button to allow these genes also to be added back to your input list. This will take back you to the previous screen, wherein after selecting the enrichment analysis parameters you can proceed with the analysis by clicking on "submit" button.
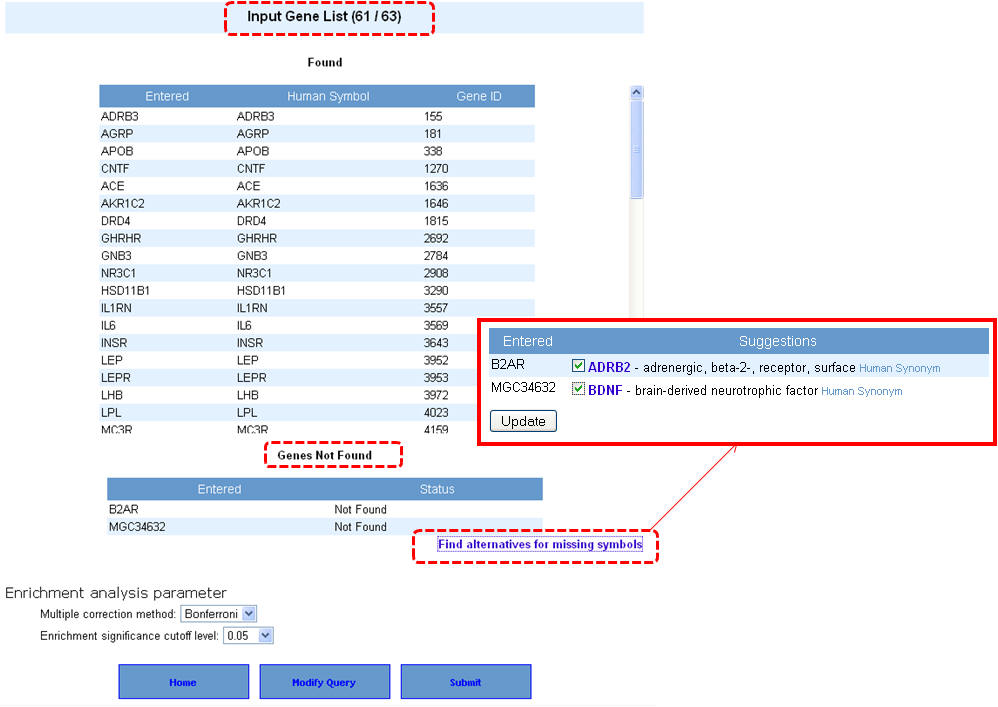
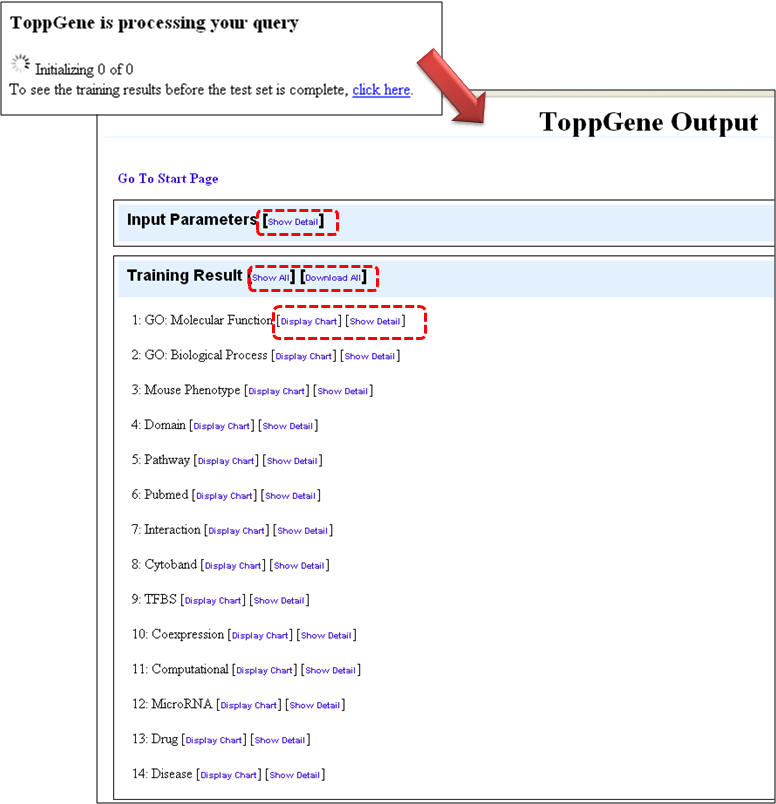
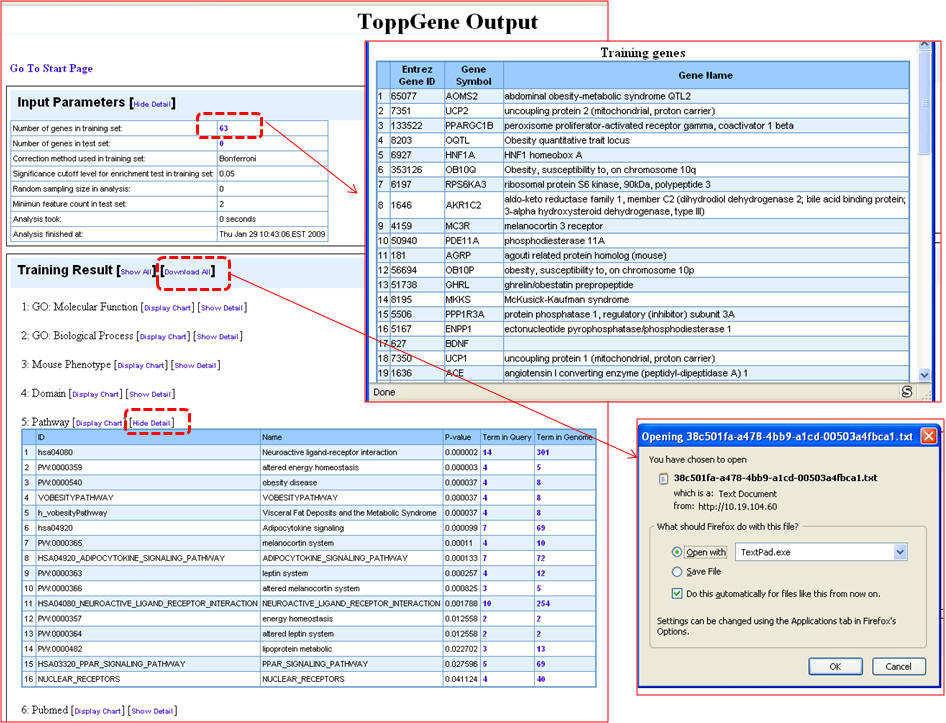
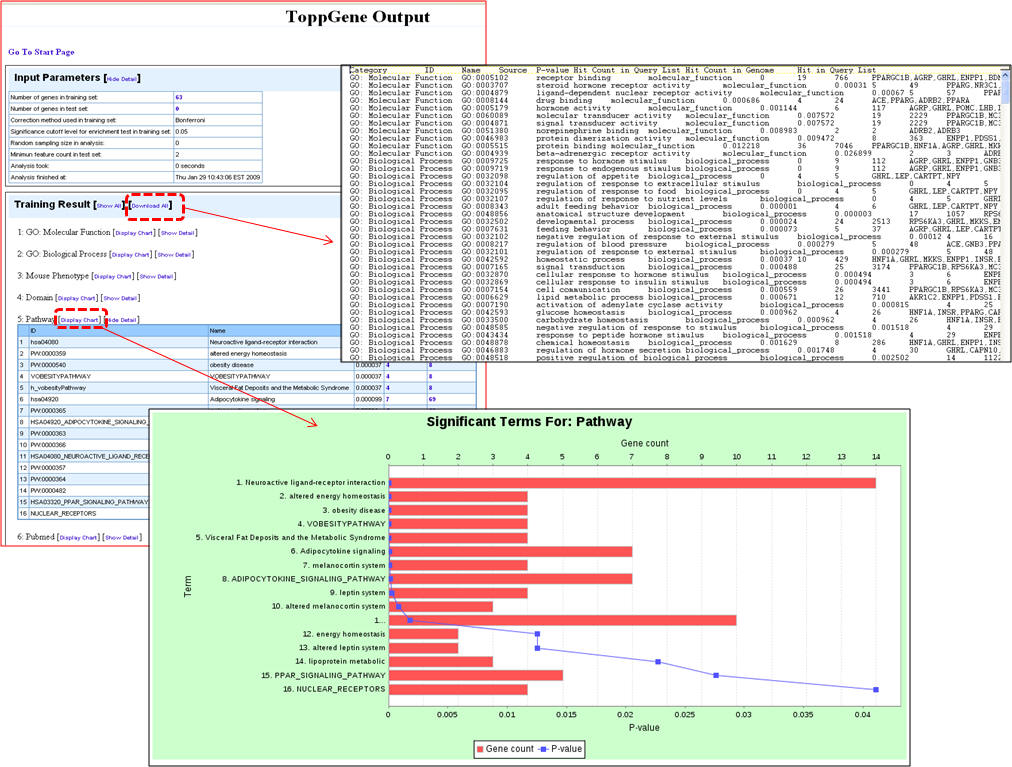
5. Once the query is processed, the summary "Toppgene Output" screen lists the input parameters and the results. Clicking on the "Show Detail" link opens a pop-up box to see the enriched terms along with p-valuesgene li
b. Using ToppGene for disease gene prioritization based on functional similarity to training set genes
Query: To rank or prioritize a list of genes (test set) by functional annotation similarity to training set.
Here, we will use a list of known obesity associated genes compiled from NCBI's OMIM and Entrez Gene as the training set. The test set is built by creating an artificial linkage interval using a GWAS identified/confirmed obesity genes. In this case, we have used MC4R gene as the candidate gene. The test set is generated by adding 99 nearest neighboring genes of MC4R on the chromosome (99+1=100 test set genes) (see Supplementary File 2 for the lists of training and test set genes) .
1. From the homepage click on the second link ("ToppGene: Candidate gene prioritization")
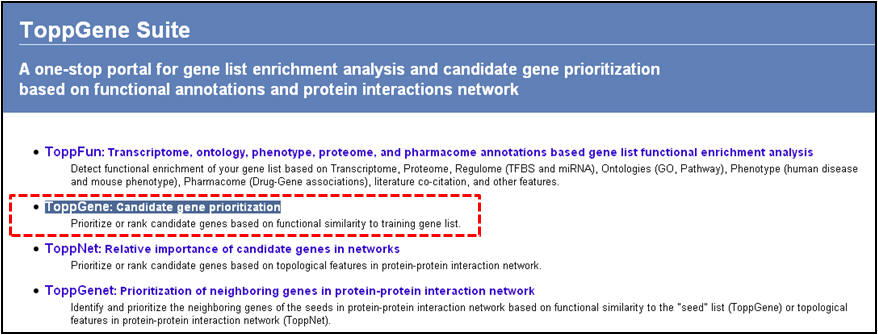
2. On the following page ("Candidate gene prioritization"), enter either gene symbols or Entrez gene IDs, and click "submit query".
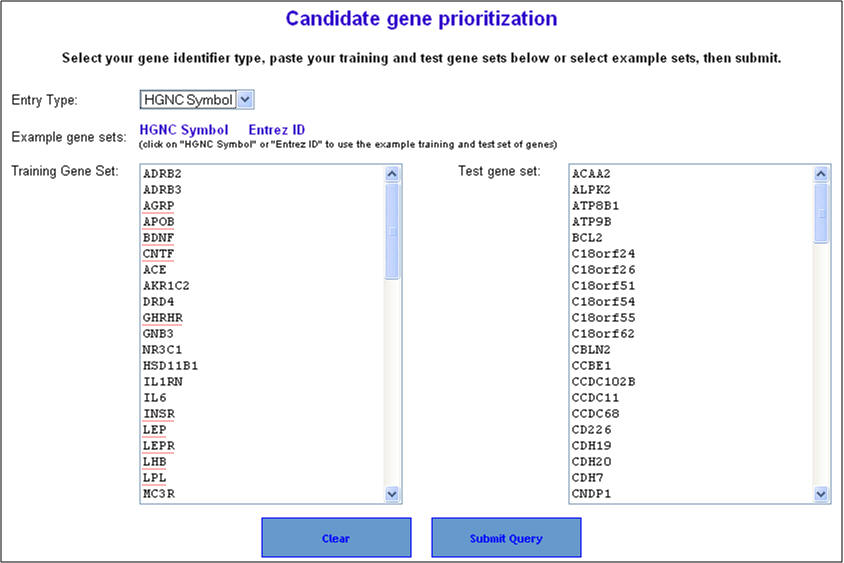
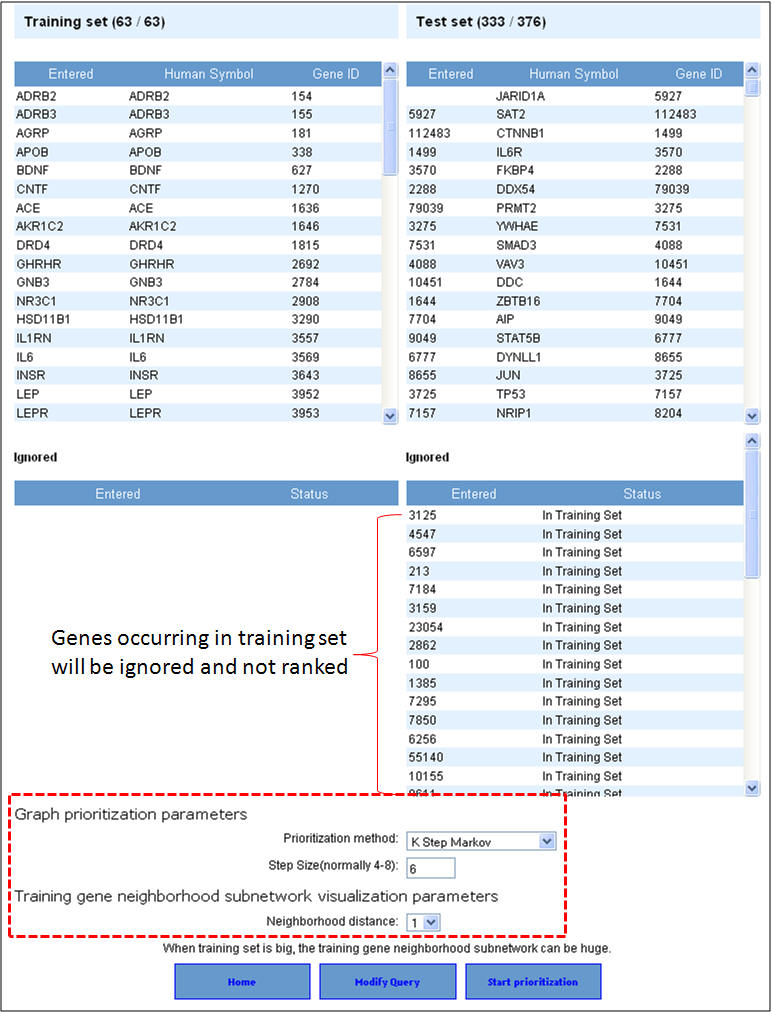
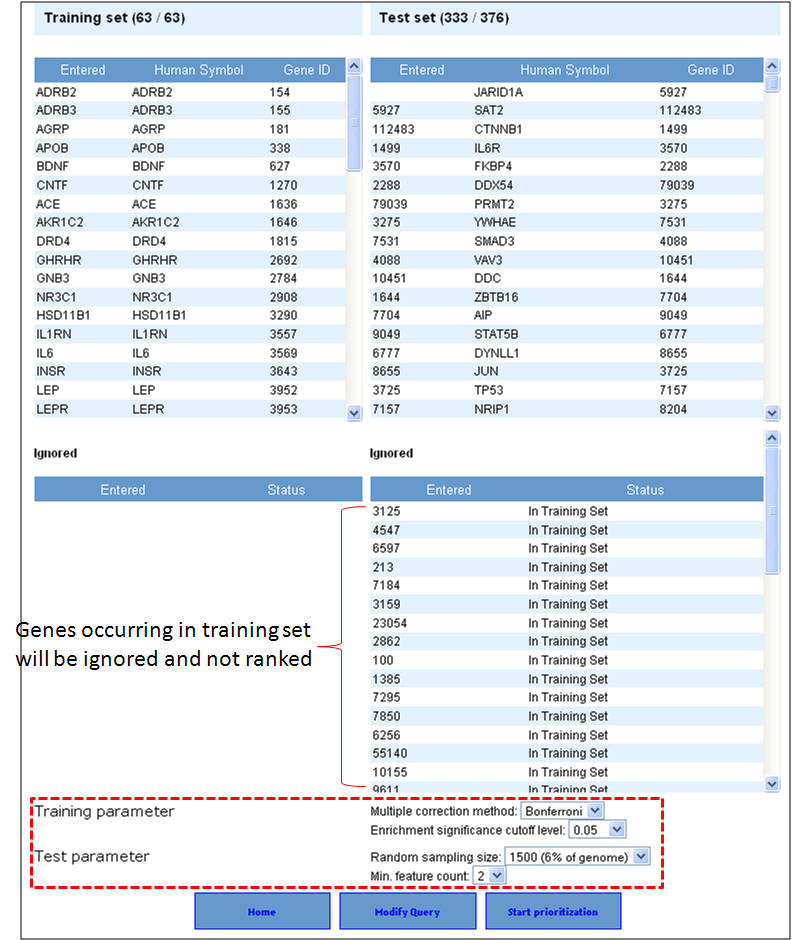
3. Select the appropriate statistical parameters. The "random sampling size" option is for selecting the background gene set from the genome for computing the p-value while the "Min. feature count" represents the number of features that need to be considered for prioritization. The default options are 6% of the genome (or 1500 genes from a total of 25000 genes) for random sampling size and feature count is 2. As described earlier in the enrichment analysis, if your gene lists contain alternate symbols or duplicates they are ignored or presented with the option to resolve them and add them back to your input list. Additionally, if there are common genes between training and test sets (i.e. test set genes which are also found in training set), these will be removed from the test set and no ranks will be assigned to them. After selecting the appropriate statistical parameters (training and test) click on the "start prioritization" button.
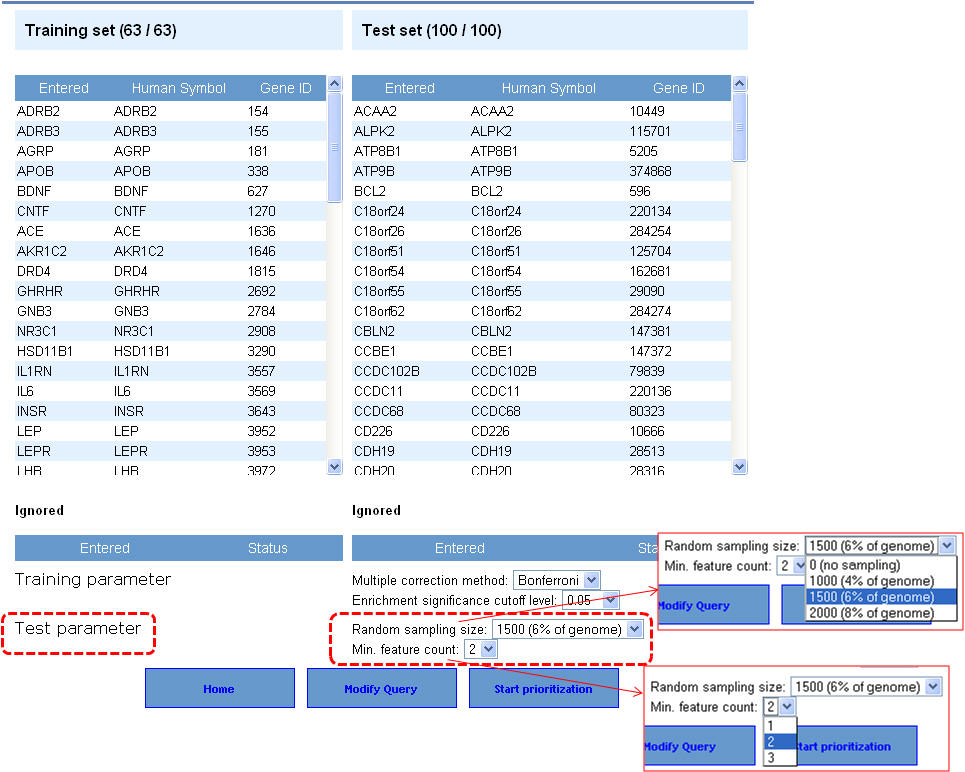
4. Once the analysis is complete, the first half of the results page is similar to what you have seen under enrichment analysis results except that the input parameters show the test set genes also. The prioritized list of test genes sorted according to their ranks based on the p-values are displayed in the lower half. Each column indicates the features used to compute similarity between training and test set.
c. Using ToppNet for disease gene prioritization based on topological features in protein-protein interactions network (PPIN)
Query: To rank or prioritize a list of genes (test set) based on topological features in PPIN.
We will again use the same list of known obesity associated genes compiled from NCBI's OMIM and Entrez Gene as the training set. The test set, as described earlier, is built by creating an artificial linkage interval using a GWAS identified/confirmed obesity genes. In this case, we have used MC4R gene as the candidate gene. The test set is generated by adding 99 nearest neighboring genes of MC4R on the chromosome (99+1=100 test set genes) (see Supplementary File 2 for the lists of training and test set genes). Both the training and test set genes are mapped onto the global PPIN and then the test set genes are scored based on how "close" they are to the training set genes.
1. From the homepage click on the third link ("ToppNet: relative importance of candidate genes in networks").
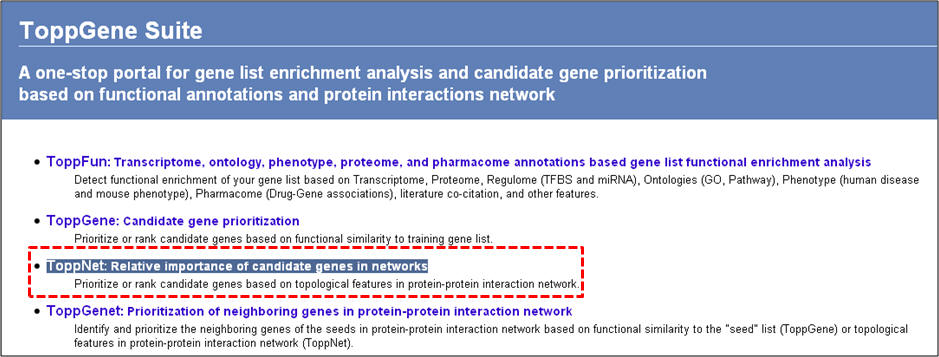
2. On the following page ("Relative importance of candidate genes in protein-protein interaction network"), enter either gene symbols or Entrez gene IDs, and click "submit query".
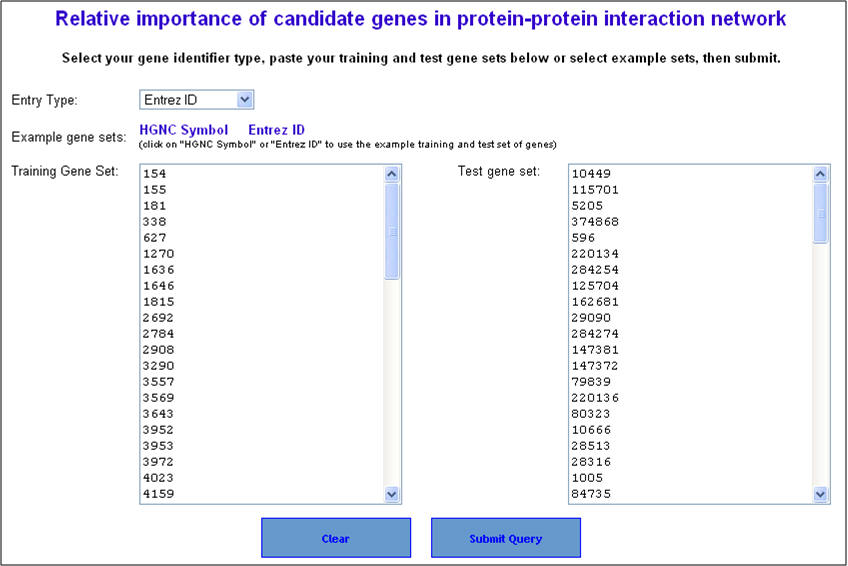
3. Select the network prioritization parameters. There are 3 options available (i) K Step Markov (default); (ii) HITS with Priors; and (iii) Page Rank with Priors. Also select the training gene neighborhood subnetwork visualization parameters (default is level 1) and then hit "Start prioritization" button.
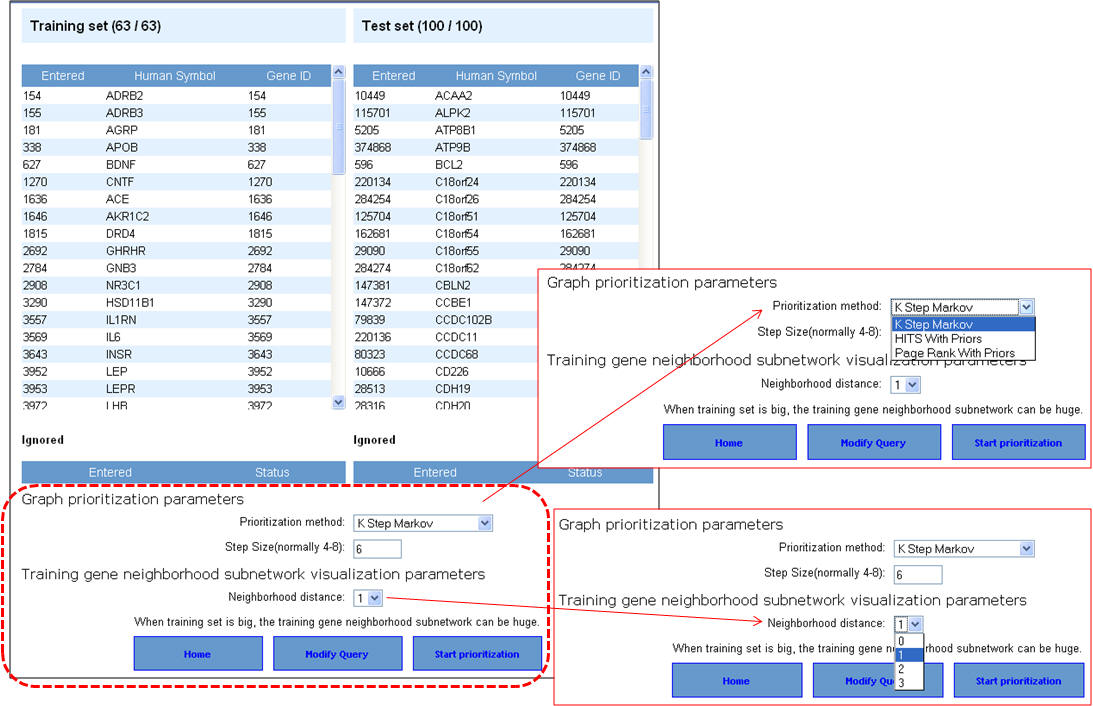
4. Following the prioritization, the test set genes are ranked according to the score. The training gene subnetwork can be exported as a Cytoscape-compatible file. Additionally, the graphical representations of the training gene subnetwork are also presented.
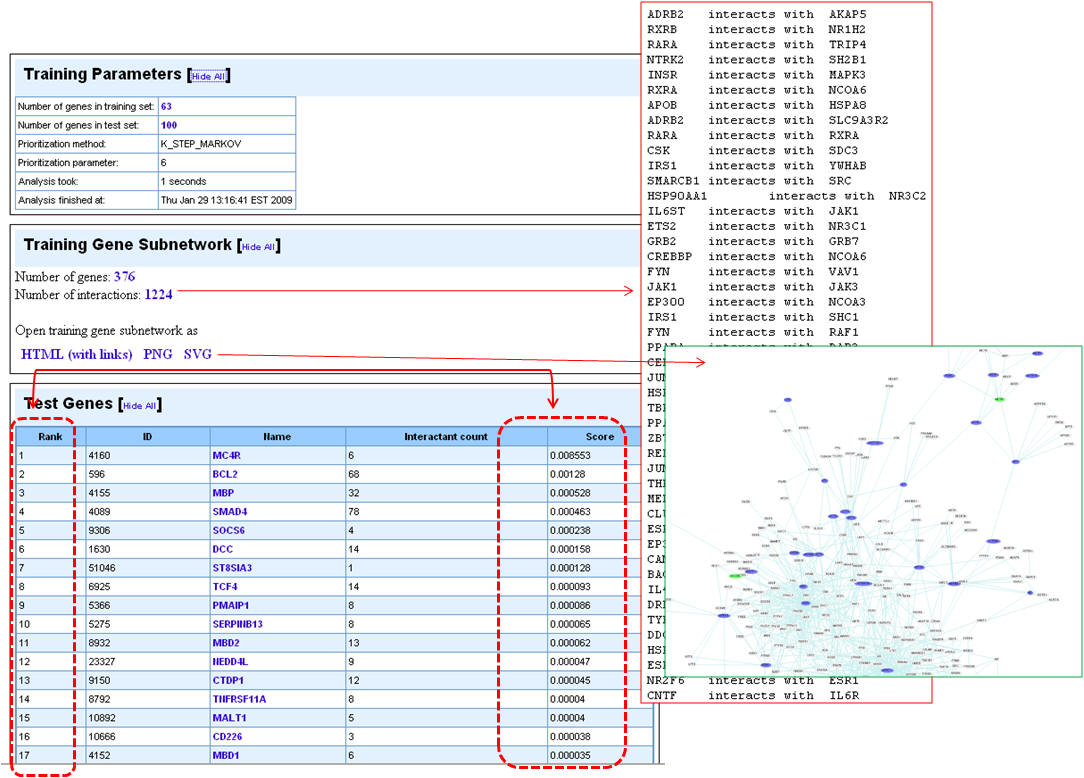
d. Using ToppGenet to identify and prioritize the neighboring genes of the "seeds" or training set in protein-protein interactions network (PPIN)
Query: To rank or prioritize a list of genes in the interactome of training set genes using either functional similarity (ToppGene) or PPIN analysis (ToppNet).
We will continue using the same list of known obesity associated genes compiled from NCBI's OMIM and Entrez Gene as the training set. The test set this time will be composed of genes that are interacting with the training set of genes (level 1 = immediate interactants).
1. From the homepage click on the fourth option ("ToppGenet: prioritization of neighboring genes in protein-protein interaction network").
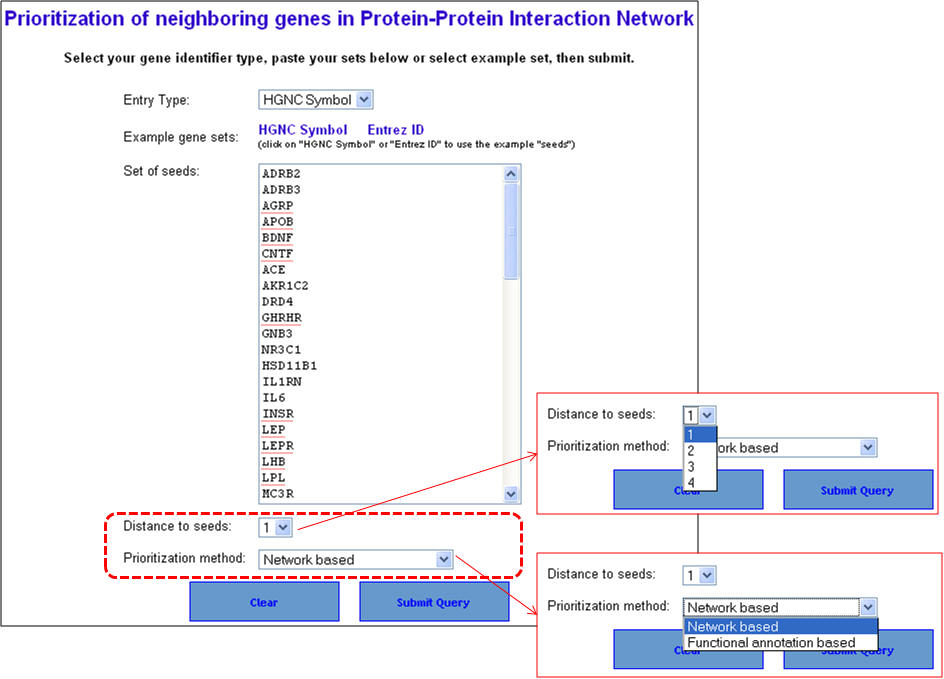
2. Enter the gene symbols and select the options as to how much interactome space you want to be considered, and whether you want to prioritization of the interactome genes to be done by functional similarity (ToppGene) or network analysis (ToppNet). When you select "distance to seeds" as 1, it means that the test set comprises all genes that are immediate interactants of the training set genes. Any overlapping or common genes between the training and test sets will be ignored from the test set and not considered for ranking.
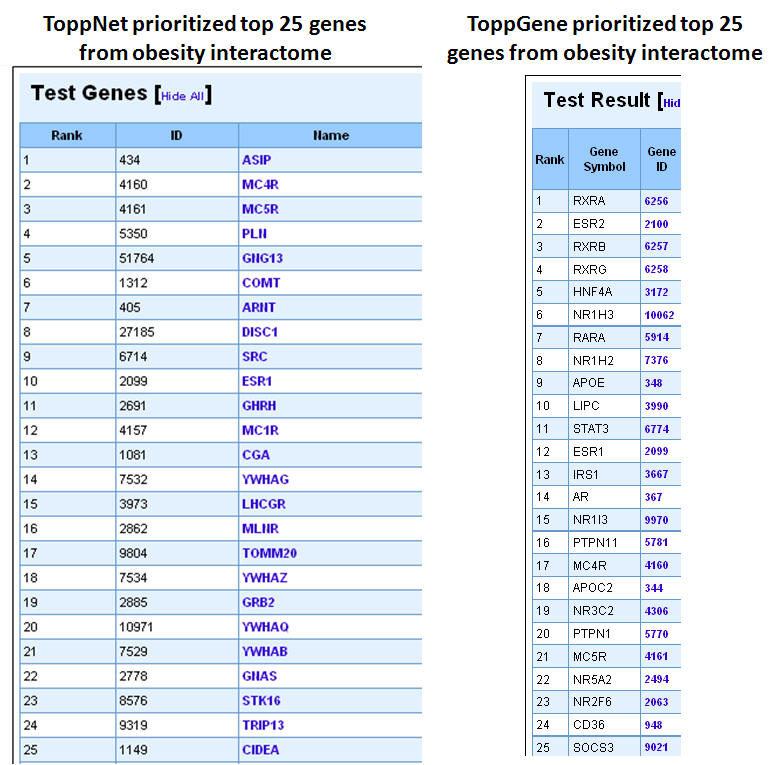
3. In this example extending the interactome to "level 1" fetches 376 genes which are used as test set after removing the common genes (i.e. those present in the training set too). The final test therefore comprises 33 genes (376-43). The first panel shows the prioritization done through PPIN analysis (ToppNet) while the lower panel shows the prioritization done based on functional similarity (ToppGene).
4. Below is a list of top 25 genes prioritized using ToppNet and ToppGene.
Supplementary File 2: List of diseases, training and test set genes used to demonstrate the utility of ToppGene and ToppNet
The training set of known disease genes were compiled from NCBI's OMIM and Entrez Gene records limiting the search option fields to "Disease/Phenotype" in the latter. The test set genes, as described earlier, were compiled by mixing the GWAS identified/confirmed candidate gene with 99 nearest neighboring genes in the genome (i.e. genes occurring in the flanking regions of the candidate gene).
i. Results of the 20 candidate gene prioritizations for the below listed five genetic diseases using ToppGene and ToppNet.
The gene-disease associations were from recently reported GWAS and include novel disease gene associations. The training sets were compiled using “phenotype/disease” annotations in NCBI’s Entrez Gene records and OMIM. To build the test set genes, we defined the artificial linkage interval to be the set of genes containing the 99 nearest neighboring genes to the novel disease gene based on their genomic distance on the same chromosome. ToppGene and ToppNet prioritization results are presented in below. ToppGene ranked 19/20 (95%) candidate genes within the top 20% while ToppNet ranked 12/16 (75%) candidate genes among the top 20%. The mean ranks for ToppGene and ToppNet based prioritization were 6.8 and 11.75 (excluding 4 disease genes that lacked interaction data) respectively.
| Disease | Reference | Gene | ToppGene Rank | ToppNet Rank |
| Bipolar Disorder | Le-Niculescu et al. | KLF12 | 2 | 15 |
| Bipolar Disorder | Le-Niculescu et al. | RORB | 4 | 18 |
| Bipolar Disorder | Le-Niculescu et al. | RORA | 7 | 13 |
| Bipolar Disorder | Le-Niculescu et al. | ALDH1A1 | 10 | No interaction data |
| Bipolar Disorder | Le-Niculescu et al. | AK3L1 | 11 | No interaction data |
| Cardiomyopathy | Dhandapany et al. | MYBPC3 | 1 | 2 |
| Celiac Disease | Hunt et al. | SH2B3 | 1 | 8 |
| Celiac Disease | Hunt et al. | CCR3 | 2 | 3 |
| Celiac Disease | Hunt et al. | IL18R1 | 3 | 29 |
| Celiac Disease | Hunt et al. | RGS1 | 9 | 26 |
| Celiac Disease | Hunt et al. | TAGAP | 14 | No interaction data |
| Celiac Disease | Hunt et al. | IL12A | 14 | 10 |
| Crohns Disease | Fisher et al. | MST1 | 1 | 27 |
| Crohns Disease | Fisher et al. | NKX2-3 | 1 | 27 |
| Crohns Disease | Fisher et al. | IRGM | 2 | No interaction data |
| Crohns Disease | Villani et al. | NLRP3 | 5 | 1 |
| Crohns Disease | Fisher et al. | IL12B | 7 | 1 |
| Crohns Disease | Barrett et al. | STAT3 | 11 | 1 |
| Franke et al. | ||||
| Crohns Disease | Franke et al. | PTPN2 | 30 | 6 |
| Obesity | Renstrom et al. | MC4R | 1 | 1 |
| Mean | 6.8 | 11.75 |
131 Known bipolar disease associated genes (Training set; compiled from OMIM and NCBI Entrez Gene)
| Entrez Gene ID | Gene Symbol | Gene Name |
| 125 | ADH1B | alcohol dehydrogenase 1B (class I), beta polypeptide |
| 207 | AKT1 | v-akt murine thymoma viral oncogene homolog 1 |
| 79796 | ALG9 | asparagine-linked glycosylation 9, alpha-1,2-mannosyltransferase homolog (S. cerevisiae) |
| 288 | ANK3 | ankyrin 3, node of Ranvier (ankyrin G) |
| 8542 | APOL1 | apolipoprotein L, 1 |
| 23780 | APOL2 | apolipoprotein L, 2 |
| 80832 | APOL4 | apolipoprotein L, 4 |
| 84100 | ARL6 | ADP-ribosylation factor-like 6 |
| 488 | ATP2A2 | ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 |
| 582 | BBS1 | Bardet-Biedl syndrome 1 |
| 79738 | BBS10 | Bardet-Biedl syndrome 10 |
| 166379 | BBS12 | Bardet-Biedl syndrome 12 |
| 583 | BBS2 | Bardet-Biedl syndrome 2 |
| 585 | BBS4 | Bardet-Biedl syndrome 4 |
| 129880 | BBS5 | Bardet-Biedl syndrome 5 |
| 55212 | BBS7 | Bardet-Biedl syndrome 7 |
| 27241 | BBS9 | Bardet-Biedl syndrome 9 |
| 627 | BDNF | brain-derived neurotrophic factor |
| 56652 | C10orf2 | chromosome 10 open reading frame 2 |
| 400359 | C15orf53 | chromosome 15 open reading frame 53 |
| 778 | CACNA1F | calcium channel, voltage-dependent, L type, alpha 1F subunit |
| 100133941 | CD24 | CD24 molecule |
| 80184 | CEP290 | centrosomal protein 290kDa |
| 1116 | CHI3L1 | chitinase 3-like 1 (cartilage glycoprotein-39) |
| 4261 | CIITA | class II, major histocompatibility complex, transactivator |
| 9575 | CLOCK | clock homolog (mouse) |
| 1243 | CMM | cutaneous malignant melanoma/dysplastic nevus |
| 474388 | CMM4 | Melanoma, cutaneous malignant, 4 |
| 1312 | COMT | catechol-O-methyltransferase |
| 1392 | CRH | corticotropin releasing hormone |
| 1610 | DAO | D-amino-acid oxidase |
| 267012 | DAOA | D-amino acid oxidase activator |
| 84516 | DCTN5 | dynactin 5 (p25) |
| 1714 | DGCR | DiGeorge syndrome chromosome region |
| 160851 | DGKH | diacylglycerol kinase, eta |
| 27185 | DISC1 | disrupted in schizophrenia 1 |
| 27184 | DISC2 | disrupted in schizophrenia 2 (non-protein coding) |
| 1756 | DMD | dystrophin |
| 1786 | DNMT1 | DNA (cytosine-5-)-methyltransferase 1 |
| 1814 | DRD3 | dopamine receptor D3 |
| 1815 | DRD4 | dopamine receptor D4 |
| 92126 | DSEL | dermatan sulfate epimerase-like |
| 84062 | DTNBP1 | dystrobrevin binding protein 1 |
| 9030 | ERDA1 | expanded repeat domain, CAG/CTG 1 |
| 2289 | FKBP5 | FK506 binding protein 5 |
| 2332 | FMR1 | fragile X mental retardation 1 |
| 282706 | G30 | protein LG30-like |
| 2555 | GABRA2 | gamma-aminobutyric acid (GABA) A receptor, alpha 2 |
| 2558 | GABRA5 | gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| 2629 | GBA | glucosidase, beta; acid (includes glucosylceramidase) |
| 2774 | GNAL | guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| 9248 | GPR50 | G protein-coupled receptor 50 |
| 2892 | GRIA3 | glutamate receptor, ionotrophic, AMPA 3 |
| 2913 | GRM3 | glutamate receptor, metabotropic 3 |
| 2932 | GSK3B | glycogen synthase kinase 3 beta |
| 3119 | HLA-DQB1 | major histocompatibility complex, class II, DQ beta 1 |
| 3123 | HLA-DRB1 | major histocompatibility complex, class II, DR beta 1 |
| 3356 | HTR2A | 5-hydroxytryptamine (serotonin) receptor 2A |
| 3359 | HTR3A | 5-hydroxytryptamine (serotonin) receptor 3A |
| 3360 | HTR4 | 5-hydroxytryptamine (serotonin) receptor 4 |
| 3575 | IL7R | interleukin 7 receptor |
| 3612 | IMPA1 | inositol(myo)-1(or 4)-monophosphatase 1 |
| 3613 | IMPA2 | inositol(myo)-1(or 4)-monophosphatase 2 |
| 3782 | KCNN3 | potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 |
| 4095 | MAFD1 | major affective disorder 1 |
| 4096 | MAFD2 | major affective disorder 2 |
| 100188792 | MAFD3 | Major affective disorder 3, early onset |
| 100126593 | MAFD4 | Major affective disorder 4 |
| 100188843 | MAFD5 | Major affective disorder 5 |
| 100188844 | MAFD6 | Major affective disorder 6 |
| 100196912 | MAFD8 | Major affective disorder-8, susceptibility to |
| 100196917 | MAFD9 | Major affective disorder-9, susceptibility to |
| 4128 | MAOA | monoamine oxidase A |
| 4157 | MC1R | melanocortin 1 receptor (alpha melanocyte stimulating hormone receptor) |
| 431708 | MDD1 | Major depressive disorder |
| 431709 | MDD2 | Major depressive disorder 2 |
| 10386 | MHW1 | Mental health wellness 1 |
| 50979 | MHW2 | Mental health wellness 2 |
| 8195 | MKKS | McKusick-Kaufman syndrome |
| 54903 | MKS1 | Meckel syndrome, type 1 |
| 378484 | MRX82 | mental retardation, X-linked 82 |
| 4397 | MS | multiple sclerosis |
| 4524 | MTHFR | 5,10-methylenetetrahydrofolate reductase (NADPH) |
| 4618 | MYF6 | myogenic factor 6 (herculin) |
| 4663 | NA | neurocanthocytosis |
| 4706 | NDUFAB1 | NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1, 8kDa |
| 23327 | NEDD4L | neural precursor cell expressed, developmentally down-regulated 4-like |
| 114548 | NLRP3 | NLR family, pyrin domain containing 3 |
| 4842 | NOS1 | nitric oxide synthase 1 (neuronal) |
| 2908 | NR3C1 | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| 60506 | NYX | nyctalopin |
| 79728 | PALB2 | partner and localizer of BRCA2 |
| 5116 | PCNT | pericentrin |
| 5158 | PDE6B | phosphodiesterase 6B, cGMP-specific, rod, beta |
| 5443 | POMC | proopiomelanocortin |
| 5573 | PRKAR1A | protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1) |
| 5788 | PTPRC | protein tyrosine phosphatase, receptor type, C |
| 10743 | RAI1 | retinoic acid induced 1 |
| 10125 | RASGRP1 | RAS guanyl releasing protein 1 (calcium and DAG-regulated) |
| 5649 | RELN | reelin |
| 5999 | RGS4 | regulator of G-protein signaling 4 |
| 6103 | RPGR | retinitis pigmentosa GTPase regulator |
| 65078 | RTN4R | reticulon 4 receptor |
| 63944 | SCZD10 | schizophrenia disorder 10 (periodic catatonia) |
| 404686 | SCZD11 | Schizophrenia susceptibility locus, chromosome 10q-related |
| 619488 | SCZD12 | schizophrenia 12 |
| 100196913 | SCZD13 | Schizophrenia, susceptibility to, 13 |
| 6378 | SCZD2 | schizophrenia disorder 2 |
| 6365 | SCZD3 | schizophrenia disorder 3 |
| 8400 | SCZD6 | schizophrenia disorder 6 |
| 8401 | SCZD7 | schizophrenia disorder 7 |
| 8806 | SCZD8 | schizophrenia disorder 8 |
| 6506 | SLC1A2 | solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| 6507 | SLC1A3 | solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| 6531 | SLC6A3 | solute carrier family 6 (neurotransmitter transporter, dopamine), member 3 |
| 6532 | SLC6A4 | solute carrier family 6 (neurotransmitter transporter, serotonin), member 4 |
| 6600 | SMCR | Smith-Magenis syndrome chromosome region |
| 6854 | SYN2 | synapsin II |
| 8867 | SYNJ1 | synaptojanin 1 |
| 134860 | TAAR9 | trace amine associated receptor 9 |
| 6899 | TBX1 | T-box 1 |
| 6925 | TCF4 | transcription factor 4 |
| 7054 | TH | tyrosine hydroxylase |
| 91147 | TMEM67 | transmembrane protein 67 |
| 121278 | TPH2 | tryptophan hydroxylase 2 |
| 22954 | TRIM32 | tripartite motif-containing 32 |
| 7226 | TRPM2 | transient receptor potential cation channel, subfamily M, member 2 |
| 123016 | TTC8 | tetratricopeptide repeat domain 8 |
| 6845 | VAMP7 | vesicle-associated membrane protein 7 |
| 7494 | XBP1 | X-box binding protein 1 |
| 7504 | XK | X-linked Kx blood group (McLeod syndrome) |
Bipolar Disorder Test Set genes
KLF12 candidate gene (KLF12 + 99 nearest neighboring genes)
| Entrez Gene ID | Gene Symbol | Gene Name |
| 87769 | A2LD1 | AIG2-like domain 1 |
| 10257 | ABCC4 | ATP-binding cassette, sub-family C (CFTR/MRP), member 4 |
| 440138 | ALG11 | asparagine-linked glycosylation 11, alpha-1,2-mannosyltransferase homolog (yeast) |
| 540 | ATP7B | ATPase, Cu++ transporting, beta polypeptide |
| 6315 | ATXN8OS | ATXN8 opposite strand (non-protein coding) |
| 93081 | C13orf27 | chromosome 13 open reading frame 27 |
| 79866 | C13orf34 | chromosome 13 open reading frame 34 |
| 440145 | C13orf37 | chromosome 13 open reading frame 37 |
| 196541 | C13orf39 | chromosome 13 open reading frame 39 |
| 83446 | CCDC70 | coiled-coil domain containing 70 |
| 26586 | CKAP2 | cytoskeleton associated protein 2 |
| 9071 | CLDN10 | claudin 10 |
| 1203 | CLN5 | ceroid-lipofuscinosis, neuronal 5 |
| 171425 | CLYBL | citrate lyase beta like |
| 170622 | COMMD6 | COMM domain containing 6 |
| 1602 | DACH1 | dachshund homolog 1 (Drosophila) |
| 1638 | DCT | dopachrome tautomerase (dopachrome delta-isomerase, tyrosine-related protein 2) |
| 79758 | DHRS12 | dehydrogenase/reductase (SDR family) member 12 |
| 81624 | DIAPH3 | diaphanous homolog 3 (Drosophila) |
| 22894 | DIS3 | DIS3 mitotic control homolog (S. cerevisiae) |
| 5611 | DNAJC3 | DnaJ (Hsp40) homolog, subfamily C, member 3 |
| 23348 | DOCK9 | dedicator of cytokinesis 9 |
| 22873 | DZIP1 | DAZ interacting protein 1 |
| 1910 | EDNRB | endothelin receptor type B |
| 220108 | FAM124A | family with sequence similarity 124A |
| 10160 | FARP1 | FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| 26224 | FBXL3 | F-box and leucine-rich repeat protein 3 |
| 2259 | FGF14 | fibroblast growth factor 14 |
| 100131561 | FKSG29 | FKSG29 |
| 122183 | FLJ40296 | FLJ40296 protein |
| 2262 | GPC5 | glypican 5 |
| 10082 | GPC6 | glypican 6 |
| 2841 | GPR18 | G protein-coupled receptor 18 |
| 160897 | GPR180 | G protein-coupled receptor 180 |
| 1880 | GPR183 | G protein-coupled receptor 183 |
| 2974 | GUCY1B2 | guanylate cyclase 1, soluble, beta 2 |
| 266722 | HS6ST3 | heparan sulfate 6-O-sulfotransferase 3 |
| 26512 | INTS6 | integrator complex subunit 6 |
| 3843 | IPO5 | importin 5 |
| 9358 | ITGBL1 | integrin, beta-like 1 (with EGF-like repeat domains) |
| 115207 | KCTD12 | potassium channel tetramerisation domain containing 12 |
| 79070 | KDELC1 | KDEL (Lys-Asp-Glu-Leu) containing 1 |
| 11278 | KLF12 | Kruppel-like factor 12 |
| 688 | KLF5 | Kruppel-like factor 5 (intestinal) |
| 57626 | KLHL1 | kelch-like 1 (Drosophila) |
| 11061 | LECT1 | leukocyte cell derived chemotaxin 1 |
| 4008 | LMO7 | LIM domain 7 |
| 79621 | RNASEH2B | ribonuclease H2, subunit B |
| 220115 | LOC220115 | TPTE pseudogene |
| 729233 | LOC729233 | FLJ40296 protein family member |
| 729240 | LOC729240 | FLJ40296 protein family member |
| 729246 | LOC729246 | FLJ40296 protein family member |
| 729250 | LOC729250 | FLJ40296 protein family member |
| 10150 | MBNL2 | muscleblind-like 2 (Drosophila) |
| 23077 | MYCBP2 | MYC binding protein 2 |
| 259232 | NALCN | sodium leak channel, non-selective |
| 54602 | NDFIP2 | Nedd4 family interacting protein 2 |
| 4752 | NEK3 | NIMA (never in mitosis gene a)-related kinase 3 |
| 341676 | NEK5 | NIMA (never in mitosis gene a)-related kinase 5 |
| 10562 | OLFM4 | olfactomedin 4 |
| 283491 | OR7E156P | olfactory receptor, family 7, subfamily E, member 156 pseudogene |
| 27199 | OXGR1 | oxoglutarate (alpha-ketoglutarate) receptor 1 |
| 5095 | PCCA | propionyl Coenzyme A carboxylase, alpha polypeptide |
| 27253 | PCDH17 | protocadherin 17 |
| 64881 | PCDH20 | protocadherin 20 |
| 5100 | PCDH8 | protocadherin 8 |
| 5101 | PCDH9 | protocadherin 9 |
| 10464 | PIBF1 | progesterone immunomodulatory binding factor 1 |
| 5457 | POU4F1 | POU class 4 homeobox 1 |
| 5911 | RAP2A | RAP2A, member of RAS oncogene family |
| 64062 | RBM26 | RNA binding motif protein 26 |
| 140432 | RNF113B | ring finger protein 113B |
| 79596 | RNF219 | ring finger protein 219 |
| 8796 | SCEL | sciellin |
| 647174 | SERPINE3 | serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 3 |
| 122060 | SLAIN1 | SLAIN motif family, member 1 |
| 6564 | SLC15A1 | solute carrier family 15 (oligopeptide transporter), member 1 |
| 114798 | SLITRK1 | SLIT and NTRK-like family, member 1 |
| 26050 | SLITRK5 | SLIT and NTRK-like family, member 5 |
| 84189 | SLITRK6 | SLIT and NTRK-like family, member 6 |
| 11166 | SOX21 | SRY (sex determining region Y)-box 21 |
| 10253 | SPRY2 | sprouty homolog 2 (Drosophila) |
| 8428 | STK24 | serine/threonine kinase 24 (STE20 homolog, yeast) |
| 10910 | SUGT1 | SGT1, suppressor of G2 allele of SKP1 (S. cerevisiae) |
| 9882 | TBC1D4 | TBC1 domain family, member 4 |
| 81550 | TDRD3 | tudor domain containing 3 |
| 23483 | TGDS | TDP-glucose 4,6-dehydratase |
| 55901 | THSD1 | thrombospondin, type I, domain containing 1 |
| 374500 | THSD1P | thrombospondin, type I, domain containing 1 pseudogene |
| 9375 | TM9SF2 | transmembrane 9 superfamily member 2 |
| 84899 | TMTC4 | transmembrane and tetratricopeptide repeat containing 4 |
| 7174 | TPP2 | tripeptidyl peptidase II |
| 337867 | UBAC2 | UBA domain containing 2 |
| 7347 | UCHL3 | ubiquitin carboxyl-terminal esterase L3 (ubiquitin thiolesterase) |
| 55757 | UGCGL2 | UDP-glucose ceramide glucosyltransferase-like 2 |
| 9724 | UTP14C | UTP14, U3 small nucleolar ribonucleoprotein, homolog C (yeast) |
| 51028 | VPS36 | vacuolar protein sorting 36 homolog (S. cerevisiae) |
| 115825 | WDFY2 | WD repeat and FYVE domain containing 2 |
| 7546 | ZIC2 | Zic family member 2 (odd-paired homolog, Drosophila) |
| 85416 | ZIC5 | Zic family member 5 (odd-paired homolog, Drosophila) |
Bipolar Disorder Test Set genes
ALDH1A1 and RORB candidate gene (ALDH1A1 and RORB + 98 nearest neighboring genes)
| Entrez Gene ID | Gene Symbol | Gene Name |
| 23287 | AGTPBP1 | ATP/GTP binding protein 1 |
| 216 | ALDH1A1 | aldehyde dehydrogenase 1 family, member A1 |
| 84210 | ANKRD20A1 | ankyrin repeat domain 20 family, member A1 |
| 441430 | ANKRD20A2 | ankyrin repeat domain 20 family, member A2 |
| 441425 | ANKRD20A3 | ankyrin repeat domain 20 family, member A3 |
| 728747 | ANKRD20A4 | ankyrin repeat domain 20 family, member A4 |
| 301 | ANXA1 | annexin A1 |
| 320 | APBA1 | amyloid beta (A4) precursor protein-binding, family A, member 1 |
| 375719 | AQP7P1 | aquaporin 7 pseudogene 1 |
| 389756 | AQP7P2 | aquaporin 7 pseudogene 2 |
| 441432 | AQP7P3 | aquaporin 7 pseudogene 3 |
| 414328 | C9orf103 | chromosome 9 open reading frame 103 |
| 138255 | C9orf135 | chromosome 9 open reading frame 135 |
| 389766 | C9orf153 | chromosome 9 open reading frame 153 |
| 55071 | C9orf40 | chromosome 9 open reading frame 40 |
| 138199 | C9orf41 | chromosome 9 open reading frame 41 |
| 138240 | C9orf57 | chromosome 9 open reading frame 57 |
| 9413 | C9orf61 | chromosome 9 open reading frame 61 |
| 84267 | C9orf64 | chromosome 9 open reading frame 64 |
| 169693 | C9orf71 | chromosome 9 open reading frame 71 |
| 286234 | C9orf79 | chromosome 9 open reading frame 79 |
| 138241 | C9orf85 | chromosome 9 open reading frame 85 |
| 54981 | C9orf95 | chromosome 9 open reading frame 95 |
| 445571 | CBWD3 | COBW domain containing 3 |
| 220869 | CBWD5 | COBW domain containing 5 |
| 644019 | CBWD6 | COBW domain containing 6 |
| 728788 | CCDC29 | coiled-coil domain containing 29 |
| 23552 | CCRK | cell cycle related kinase |
| 84131 | CEP78 | centrosomal protein 78kDa |
| 645345 | CHCHD9 | coiled-coil-helix-coiled-coil-helix domain containing 9 |
| 1514 | CTSL1 | cathepsin L1 |
| 392360 | CTSL3 | cathepsin L family member 3 |
| 1612 | DAPK1 | death-associated protein kinase 1 |
| 51104 | FAM108B1 | family with sequence similarity 108, member B1 |
| 116224 | FAM122A | family with sequence similarity 122A |
| 548321 | FAM27A | family with sequence similarity 27, member A |
| 727905 | FAM75A5 | family with sequence similarity 75, member A5 |
| 26165 | FAM75A7 | family with sequence similarity 75, member A7 |
| 404770 | FAM75B | family with sequence similarity 75, member B |
| 401535 | FLJ45537 | FLJ45537 protein |
| 389763 | FLJ46321 | FLJ46321 protein |
| 442425 | FOXB2 | forkhead box B2 |
| 100036519 | FOXD4L2 | forkhead box D4-like 2 |
| 286380 | FOXD4L3 | forkhead box D4-like 3 |
| 349334 | FOXD4L4 | forkhead box D4-like 4 |
| 653427 | FOXD4L5 | forkhead box D4-like 5 |
| 653404 | FOXD4L6 | forkhead box D4-like 6 |
| 257019 | FRMD3 | FERM domain containing 3 |
| 2395 | FXN | frataxin |
| 2619 | GAS1 | growth arrest-specific 1 |
| 2650 | GCNT1 | glucosaminyl (N-acetyl) transferase 1, core 2 (beta-1,6-N-acetylglucosaminyltransferase) |
| 9615 | GDA | guanine deaminase |
| 80318 | GKAP1 | G kinase anchoring protein 1 |
| 9630 | GNA14 | guanine nucleotide binding protein (G protein), alpha 14 |
| 2776 | GNAQ | guanine nucleotide binding protein (G protein), q polypeptide |
| 51280 | GOLM1 | golgi membrane protein 1 |
| 3190 | HNRNPK | heterogeneous nuclear ribonucleoprotein K |
| 653553 | HSPBL2 | heat shock 27kDa protein-like 2 pseudogene |
| 81689 | ISCA1 | iron-sulfur cluster assembly 1 homolog (S. cerevisiae) |
| 387628 | KGFLP1 | keratinocyte growth factor-like protein 1 |
| 654466 | KGFLP2 | keratinocyte growth factor-like protein 2 |
| 55582 | KIF27 | kinesin family member 27 |
| 687 | KLF9 | Kruppel-like factor 9 |
| 100133920 | LOC100133920 | hypothetical protein LOC100133920 |
| 440896 | LOC440896 | hypothetical gene LOC440896 |
| 442421 | LOC442421 | hypothetical LOC442421 |
| 572558 | LOC572558 | hypothetical locus LOC572558 |
| 60560 | MAK10 | MAK10 homolog, amino-acid N-acetyltransferase subunit (S. cerevisiae) |
| 256691 | MAMDC2 | MAM domain containing 2 |
| 389741 | MGC21881 | hypothetical locus MGC21881 |
| 4915 | NTRK2 | neurotrophic tyrosine kinase, receptor, type 2 |
| 158046 | NXNL2 | nucleoredoxin-like 2 |
| 26578 | OSTF1 | osteoclast stimulating factor 1 |
| 50652 | PCA3 | prostate cancer antigen 3 (non-protein coding) |
| 5125 | PCSK5 | proprotein convertase subtilisin/kexin type 5 |
| 5239 | PGM5 | phosphoglucomutase 5 |
| 595135 | PGM5P2 | phosphoglucomutase 5 pseudogene 2 |
| 8395 | PIP5K1B | phosphatidylinositol-4-phosphate 5-kinase, type I, beta |
| 5568 | PRKACG | protein kinase, cAMP-dependent, catalytic, gamma |
| 158471 | PRUNE2 | prune homolog 2 (Drosophila) |
| 29968 | PSAT1 | phosphoserine aminotransferase 1 |
| 375743 | PTAR1 | protein prenyltransferase alpha subunit repeat containing 1 |
| 158158 | RASEF | RAS and EF-hand domain containing |
| 55312 | RFK | riboflavin kinase |
| 80010 | RMI1 | RMI1, RecQ mediated genome instability 1, homolog (S. cerevisiae) |
| 6096 | RORB | RAR-related orphan receptor B |
| 64078 | SLC28A3 | solute carrier family 28 (sodium-coupled nucleoside transporter), member 3 |
| 23137 | SMC5 | structural maintenance of chromosomes 5 |
| 10927 | SPIN1 | spindlin 1 |
| 9414 | TJP2 | tight junction protein 2 (zona occludens 2) |
| 7088 | TLE1 | transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila) |
| 7091 | TLE4 | transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| 117531 | TMC1 | transmembrane channel-like 1 |
| 23670 | TMEM2 | transmembrane protein 2 |
| 80036 | TRPM3 | transient receptor potential cation channel, subfamily M, member 3 |
| 140803 | TRPM6 | transient receptor potential cation channel, subfamily M, member 6 |
| 29979 | UBQLN1 | ubiquilin 1 |
| 23230 | VPS13A | vacuolar protein sorting 13 homolog A (S. cerevisiae) |
| 79670 | ZCCHC6 | zinc finger, CCHC domain containing 6 |
| 7763 | ZFAND5 | zinc finger, AN1-type domain 5 |
Bipolar Disorder Test Set genes
RORA candidate gene (RORA + 99 nearest neighboring genes)
| Entrez Gene ID | Gene Symbol | Gene Name |
| 102 | ADAM10 | ADAM metallopeptidase domain 10 |
| 8854 | ALDH1A2 | aldehyde dehydrogenase 1 family, member A2 |
| 348094 | ANKDD1A | ankyrin repeat and death domain containing 1A |
| 302 | ANXA2 | annexin A2 |
| 83464 | APH1B | anterior pharynx defective 1 homolog B (C. elegans) |
| 366 | AQP9 | aquaporin 9 |
| 10776 | ARPP-19 | cyclic AMP phosphoprotein, 19 kD |
| 10017 | BCL2L10 | BCL2-like 10 (apoptosis facilitator) |
| 663 | BNIP2 | BCL2/adenovirus E1B 19kDa interacting protein 2 |
| 51187 | C15orf15 | chromosome 15 open reading frame 15 |
| 81556 | C15orf44 | chromosome 15 open reading frame 44 |
| 771 | CA12 | carbonic anhydrase XII |
| 9133 | CCNB2 | cyclin B2 |
| 9236 | CCPG1 | cell cycle progression 1 |
| 84952 | CGNL1 | cingulin-like 1 |
| 8483 | CILP | cartilage intermediate layer protein, nucleotide pyrophosphohydrolase |
| 10845 | CLPX | ClpX caseinolytic peptidase X homolog (E. coli) |
| 53944 | CSNK1G1 | casein kinase 1, gamma 1 |
| 23604 | DAPK2 | death-associated protein kinase 2 |
| 10260 | DENND4A | DENN/MADD domain containing 4A |
| 115752 | DIS3L | DIS3 mitotic control homolog (S. cerevisiae)-like |
| 54878 | DPP8 | dipeptidyl-peptidase 8 |
| 161582 | DYX1C1 | dyslexia susceptibility 1 candidate 1 |
| 145741 | FAM148A | family with sequence similarity 148, member A |
| 388125 | FAM148B | family with sequence similarity 148, member B |
| 54629 | FAM63B | family with sequence similarity 63, member B |
| 145773 | FAM81A | family with sequence similarity 81, member A |
| 84191 | FAM96A | family with sequence similarity 96, member A |
| 283807 | FBXL22 | F-box and leucine-rich repeat protein 22 |
| 27023 | FOXB1 | forkhead box B1 |
| 9245 | GCNT3 | glucosaminyl (N-acetyl) transferase 3, mucin type |
| 145781 | GCOM1 | GRINL1A complex locus |
| 10681 | GNB5 | guanine nucleotide binding protein (G protein), beta 5 |
| 81488 | GRINL1A | glutamate receptor, ionotropic, N-methyl D-aspartate-like 1A |
| 2958 | GTF2A2 | general transcription factor IIA, 2, 12kDa |
| 390594 | hCG_1645727 | hCG1645727 |
| 8925 | HERC1 | hect (homologous to the E6-AP (UBE3A) carboxyl terminus) domain and RCC1 (CHC1)-like domain (RLD) 1 |
| 664618 | HSP90AB4P | heat shock protein 90kDa alpha (cytosolic), class B member 4 (pseudogene) |
| 9543 | IGDCC3 | immunoglobulin superfamily, DCC subclass, member 3 |
| 57722 | IGDCC4 | immunoglobulin superfamily, DCC subclass, member 4 |
| 9768 | KIAA0101 | KIAA0101 |
| 56204 | KIAA1370 | KIAA1370 |
| 114294 | LACTB | lactamase, beta |
| 92483 | LDHAL6B | lactate dehydrogenase A-like 6B |
| 123169 | LEO1 | Leo1, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae) |
| 3990 | LIPC | lipase, hepatic |
| 145783 | LOC145783 | hypothetical LOC145783 |
| 283663 | LOC283663 | hypothetical LOC283663 |
| 5604 | MAP2K1 | mitogen-activated protein kinase kinase 1 |
| 5597 | MAPK6 | mitogen-activated protein kinase 6 |
| 84465 | MEGF11 | multiple EGF-like-domains 11 |
| 55329 | MNS1 | meiosis-specific nuclear structural 1 |
| 123263 | MTFMT | mitochondrial methionyl-tRNA formyltransferase |
| 4643 | MYO1E | myosin IE |
| 4644 | MYO5A | myosin VA (heavy chain 12, myoxin) |
| 55930 | MYO5C | myosin VC |
| 79664 | NARG2 | NMDA receptor regulated 2 |
| 4734 | NEDD4 | neural precursor cell expressed, developmentally down-regulated 4 |
| 4947 | OAZ2 | ornithine decarboxylase antizyme 2 |
| 3175 | ONECUT1 | one cut homeobox 1 |
| 123264 | OSTbeta | organic solute transporter beta |
| 54956 | PARP16 | poly (ADP-ribose) polymerase family, member 16 |
| 10081 | PDCD7 | programmed cell death 7 |
| 80119 | PIF1 | PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| 9488 | PIGB | phosphatidylinositol glycan anchor biosynthesis, class B |
| 80301 | PLEKHO2 | pleckstrin homology domain containing, family O member 2 |
| 5479 | PPIB | peptidylprolyl isomerase B (cyclophilin B) |
| 283659 | PRTG | protogenin homolog (Gallus gallus) |
| 51495 | PTPLAD1 | protein tyrosine phosphatase-like A domain containing 1 |
| 26108 | PYGO1 | pygopus homolog 1 (Drosophila) |
| 8766 | RAB11A | RAB11A, member RAS oncogene family |
| 5873 | RAB27A | RAB27A, member RAS oncogene family |
| 51762 | RAB8B | RAB8B, member RAS oncogene family |
| 51285 | RASL12 | RAS-like, family 12 |
| 348093 | RBPMS2 | RNA binding protein with multiple splicing 2 |
| 64864 | RFX7 | regulatory factor X, 7 |
| 54778 | RNF111 | ring finger protein 111 |
| 6095 | RORA | RAR-related orphan receptor A |
| 6124 | RPL4 | ribosomal protein L4 |
| 51065 | RPS27L | ribosomal protein S27-like |
| 692149 | SCARNA14 | small Cajal body-specific RNA 14 |
| 9187 | SLC24A1 | solute carrier family 24 (sodium/potassium/calcium exchanger), member 1 |
| 79811 | SLTM | SAFB-like, transcription modulator |
| 10302 | SNAPC5 | small nuclear RNA activating complex, polypeptide 5, 19kDa |
| 6642 | SNX1 | sorting nexin 1 |
| 79856 | SNX22 | sorting nexin 22 |
| 51324 | SPG21 | spastic paraplegia 21 (autosomal recessive, Mast syndrome) |
| 6938 | TCF12 | transcription factor 12 |
| 374618 | TEX9 | testis expressed 9 |
| 54962 | TIPIN | TIMELESS interacting protein |
| 83660 | TLN2 | talin 2 |
| 29766 | TMOD3 | tropomodulin 3 (ubiquitous) |
| 7168 | TPM1 | tropomyosin 1 (alpha) |
| 9325 | TRIP4 | thyroid hormone receptor interactor 4 |
| 440279 | UNC13C | unc-13 homolog C (C. elegans) |
| 9960 | USP3 | ubiquitin specific peptidase 3 |
| 54832 | VPS13C | vacuolar protein sorting 13 homolog C (S. cerevisiae) |
| 256764 | WDR72 | WD repeat domain 72 |
| 54816 | ZNF280D | zinc finger protein 280D |
| 23060 | ZNF609 | zinc finger protein 609 |
Bipolar Disorder Test Set genes
AK3L1 candidate gene (AK3L1 + 99 nearest neighboring genes)
| Entrez Gene ID | Gene Symbol | Gene Name |
| 34 | ACADM | acyl-Coenzyme A dehydrogenase, C-4 to C-12 straight chain |
| 26027 | ACOT11 | acyl-CoA thioesterase 11 |
| 205 | AK3L1 | adenylate kinase 3-like 1 |
| 29929 | ALG6 | asparagine-linked glycosylation 6, alpha-1,3-glucosyltransferase homolog (S. cerevisiae) |
| 27329 | ANGPTL3 | angiopoietin-like 3 |
| 81573 | ANKRD13C | ankyrin repeat domain 13C |
| 84938 | ATG4C | ATG4 autophagy related 4 homolog C (S. cerevisiae) |
| 7809 | BSND | Bartter syndrome, infantile, with sensorineural deafness (Barttin) |
| 400757 | C1orf141 | chromosome 1 open reading frame 141 |
| 199920 | C1orf168 | chromosome 1 open reading frame 168 |
| 127254 | C1orf173 | chromosome 1 open reading frame 173 |
| 374977 | C1orf175 | chromosome 1 open reading frame 175 |
| 163747 | C1orf177 | chromosome 1 open reading frame 177 |
| 127428 | C1orf83 | chromosome 1 open reading frame 83 |
| 127795 | C1orf87 | chromosome 1 open reading frame 87 |
| 731 | C8A | complement component 8, alpha polypeptide |
| 732 | C8B | complement component 8, beta polypeptide |
| 57685 | CACHD1 | cache domain containing 1 |
| 200008 | CDCP2 | CUB domain containing protein 2 |
| 1429 | CRYZ | crystallin, zeta (quinone reductase) |
| 1491 | CTH | cystathionase (cystathionine gamma-lyase) |
| 606495 | CYB5RL | cytochrome b5 reductase-like |
| 1573 | CYP2J2 | cytochrome P450, family 2, subfamily J, polypeptide 2 |
| 1600 | DAB1 | disabled homolog 1 (Drosophila) |
| 55635 | DEPDC1 | DEP domain containing 1 |
| 1718 | DHCR24 | 24-dehydrocholesterol reductase |
| 1733 | DIO1 | deiodinase, iodothyronine, type I |
| 9077 | DIRAS3 | DIRAS family, GTP-binding RAS-like 3 |
| 79469 | DLEU2L | deleted in lymphocytic leukemia 2-like |
| 9829 | DNAJC6 | DnaJ (Hsp40) homolog, subfamily C, member 6 |
| 85440 | DOCK7 | dedicator of cytokinesis 7 |
| 84455 | EFCAB7 | EF-hand calcium binding domain 7 |
| 338094 | FAM151A | family with sequence similarity 151, member A |
| 55277 | FGGY | FGGY carbohydrate kinase domain containing |
| 27022 | FOXD3 | forkhead box D3 |
| 8790 | FPGT | fucose-1-phosphate guanylyltransferase |
| 1647 | GADD45A | growth arrest and DNA-damage-inducible, alpha |
| 148979 | GLIS1 | GLIS family zinc finger 1 |
| 55970 | GNG12 | guanine nucleotide binding protein (G protein), gamma 12 |
| 79971 | GPR177 | G protein-coupled receptor 177 |
| 11147 | HHLA3 | HERV-H LTR-associating 3 |
| 51361 | HOOK1 | hook homolog 1 (Drosophila) |
| 51668 | HSPB11 | heat shock protein family B (small), member 11 |
| 3595 | IL12RB2 | interleukin 12 receptor, beta 2 |
| 149233 | IL23R | interleukin 23 receptor |
| 10207 | INADL | InaD-like (Drosophila) |
| 10022 | INSL5 | insulin-like 5 |
| 23421 | ITGB3BP | integrin beta 3 binding protein (beta3-endonexin) |
| 3716 | JAK1 | Janus kinase 1 (a protein tyrosine kinase) |
| 3725 | JUN | jun oncogene |
| 163782 | KANK4 | KN motif and ankyrin repeat domains 4 |
| 54596 | L1TD1 | LINE-1 type transposase domain containing 1 |
| 388633 | LDLRAD1 | low density lipoprotein receptor class A domain containing 1 |
| 3953 | LEPR | leptin receptor |
| 54741 | LEPROT | leptin receptor overlapping transcript |
| 431707 | LHX8 | LIM homeobox 8 |
| 55631 | LRRC40 | leucine rich repeat containing 40 |
| 115353 | LRRC42 | leucine rich repeat containing 42 |
| 57554 | LRRC7 | leucine rich repeat containing 7 |
| 127255 | LRRIQ3 | leucine-rich repeats and IQ motif containing 3 |
| 57708 | MIER1 | mesoderm induction early response 1 homolog (Xenopus laevis) |
| 51253 | MRPL37 | mitochondrial ribosomal protein L37 |
| 114803 | MYSM1 | myb-like, SWIRM and MPN domains 1 |
| 257194 | NEGR1 | neuronal growth regulator 1 |
| 4774 | NFIA | nuclear factor I/A |
| 115209 | OMA1 | OMA1 homolog, zinc metallopeptidase (S. cerevisiae) |
| 25973 | PARS2 | prolyl-tRNA synthetase 2, mitochondrial (putative) |
| 255738 | PCSK9 | proprotein convertase subtilisin/kexin type 9 |
| 5142 | PDE4B | phosphodiesterase 4B, cAMP-specific (phosphodiesterase E4 dunce homolog, Drosophila) |
| 55276 | PGM2 | phosphoglucomutase 2 |
| 5301 | PIN1L | peptidylprolyl cis/trans isomerase, NIMA-interacting 1-like (pseudogene) |
| 8613 | PPAP2B | phosphatidic acid phosphatase type 2B |
| 5563 | PRKAA2 | protein kinase, AMP-activated, alpha 2 catalytic subunit |
| 5733 | PTGER3 | prostaglandin E receptor 3 (subtype EP3) |
| 55225 | RAVER2 | ribonucleoprotein, PTB-binding 2 |
| 4919 | ROR1 | receptor tyrosine kinase-like orphan receptor 1 |
| 6121 | RPE65 | retinal pigment epithelium-specific protein 65kDa |
| 26135 | SERBP1 | SERPINE1 mRNA binding protein 1 |
| 9295 | SFRS11 | splicing factor, arginine/serine-rich 11 |
| 84251 | SGIP1 | SH3-domain GRB2-like (endophilin) interacting protein 1 |
| 23169 | SLC35D1 | solute carrier family 35 (UDP-glucuronic acid/UDP-N-acetylgalactosamine dual transporter), member D1 |
| 204962 | SLC44A5 | solute carrier family 44, member 5 |
| 23648 | SSBP3 | single stranded DNA binding protein 3 |
| 4070 | TACSTD2 | tumor-associated calcium signal transducer 2 |
| 200132 | TCTEX1D1 | Tctex1 domain containing 1 |
| 83941 | TM2D1 | TM2 domain containing 1 |
| 55706 | TMEM48 | transmembrane protein 48 |
| 9528 | TMEM59 | transmembrane protein 59 |
| 199964 | TMEM61 | transmembrane protein 61 |
| 51086 | TNNI3K | TNNI3 interacting kinase |
| 55001 | TTC22 | tetratricopeptide repeat domain 22 |
| 7268 | TTC4 | tetratricopeptide repeat domain 4 |
| 127253 | TYW3 | tRNA-yW synthesizing protein 3 homolog (S. cerevisiae) |
| 148581 | UBE2U | ubiquitin-conjugating enzyme E2U (putative) |
| 7398 | USP1 | ubiquitin specific peptidase 1 |
| 23358 | USP24 | ubiquitin specific peptidase 24 |
| 79819 | WDR78 | WD repeat domain 78 |
| 54432 | YIPF1 | Yip1 domain family, member 1 |
| 9406 | ZRANB2 | zinc finger, RAN-binding domain containing 2 |
| 644928 | hCG_2033311 | hCG2033311 |
98 Known cardiomyopathy associated genes (Training set; compiled from OMIM and NCBI Entrez Gene)
| Entrez Gene ID | Gene Symbol | Gene Name |
| 37 | ACADVL | acyl-Coenzyme A dehydrogenase, very long chain |
| 58 | ACTA1 | actin, alpha 1, skeletal muscle |
| 70 | ACTC1 | actin, alpha, cardiac muscle 1 |
| 88 | ACTN2 | actinin, alpha 2 |
| 178 | AGL | amylo-1, 6-glucosidase, 4-alpha-glucanotransferase |
| 186 | AGTR2 | angiotensin II receptor, type 2 |
| 335 | APOA1 | apolipoprotein A-I |
| 788 | SLC25A20 | solute carrier family 25 (carnitine/acylcarnitine translocase), member 20 |
| 859 | CAV3 | caveolin 3 |
| 1218 | CMD1B | cardiomyopathy, dilated 1B (autosomal dominant) |
| 1219 | CMD1C | cardiomyopathy, dilated 1C (autosomal dominant) |
| 1222 | CMD1F | cardiomyopathy, dilated 1F (autosomal dominant) |
| 1224 | CMD2A | cardiomyopathy, dilated 2A (autosomal recessive) |
| 1245 | CMR1A | cardiomyopathy, restrictive 1A (autosomal dominant) |
| 1246 | CMR2A | cardiomyopathy, restrictive 2A (autosomal recessive) |
| 1247 | CMR3A | cardiomyopathy, restrictive 3A (X-linked) |
| 1352 | COX10 | COX10 homolog, cytochrome c oxidase assembly protein, heme A: farnesyltransferase (yeast) |
| 1355 | COX15 | COX15 homolog, cytochrome c oxidase assembly protein (yeast) |
| 1584 | CYP11B1 | cytochrome P450, family 11, subfamily B, polypeptide 1 |
| 1674 | DES | desmin |
| 1756 | DMD | dystrophin |
| 1824 | DSC2 | desmocollin 2 |
| 1829 | DSG2 | desmoglein 2 |
| 1832 | DSP | desmoplakin |
| 2070 | EYA4 | eyes absent homolog 4 (Drosophila) |
| 2218 | FKTN | fukutin |
| 2243 | FGA | fibrinogen alpha chain |
| 2273 | FHL1 | four and a half LIM domains 1 |
| 2395 | FXN | frataxin |
| 2632 | GBE1 | glucan (1,4-alpha-), branching enzyme 1 |
| 2706 | GJB2 | gap junction protein, beta 2, 26kDa |
| 2717 | GLA | galactosidase, alpha |
| 2720 | GLB1 | galactosidase, beta 1 |
| 2934 | GSN | gelsolin (amyloidosis, Finnish type) |
| 3030 | HADHA | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), alpha subunit |
| 3265 | HRAS | v-Ha-ras Harvey rat sarcoma viral oncogene homolog |
| 3425 | IDUA | iduronidase, alpha-L- |
| 3728 | JUP | junction plakoglobin |
| 3845 | KRAS | v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog |
| 3920 | LAMP2 | lysosomal-associated membrane protein 2 |
| 4000 | LMNA | lamin A/C |
| 4624 | MYH6 | myosin, heavy chain 6, cardiac muscle, alpha |
| 4625 | MYH7 | myosin, heavy chain 7, cardiac muscle, beta |
| 4633 | MYL2 | myosin, light chain 2, regulatory, cardiac, slow |
| 4634 | MYL3 | myosin, light chain 3, alkali; ventricular, skeletal, slow |
| 4646 | MYO6 | myosin VI |
| 4695 | NDUFA2 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2, 8kDa |
| 4720 | NDUFS2 | NADH dehydrogenase (ubiquinone) Fe-S protein 2, 49kDa (NADH-coenzyme Q reductase) |
| 4728 | NDUFS8 | NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| 4729 | NDUFV2 | NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa |
| 5250 | SLC25A3 | solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3 |
| 5318 | PKP2 | plakophilin 2 |
| 5350 | PLN | phospholamban |
| 5428 | POLG | polymerase (DNA directed), gamma |
| 5663 | PSEN1 | presenilin 1 |
| 5664 | PSEN2 | presenilin 2 (Alzheimer disease 4) |
| 5894 | RAF1 | v-raf-1 murine leukemia viral oncogene homolog 1 |
| 6197 | RPS6KA3 | ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| 6331 | SCN5A | sodium channel, voltage-gated, type V, alpha subunit |
| 6443 | SGCB | sarcoglycan, beta (43kDa dystrophin-associated glycoprotein) |
| 6444 | SGCD | sarcoglycan, delta (35kDa dystrophin-associated glycoprotein) |
| 6445 | SGCG | sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein) |
| 6584 | SLC22A5 | solute carrier family 22 (organic cation/carnitine transporter), member 5 |
| 6648 | SOD2 | superoxide dismutase 2, mitochondrial |
| 6654 | SOS1 | son of sevenless homolog 1 (Drosophila) |
| 6901 | TAZ | tafazzin |
| 7112 | TMPO | thymopoietin |
| 7134 | TNNC1 | troponin C type 1 (slow) |
| 7137 | TNNI3 | troponin I type 3 (cardiac) |
| 7139 | TNNT2 | troponin T type 2 (cardiac) |
| 7140 | TNNT3 | troponin T type 3 (skeletal, fast) |
| 7168 | TPM1 | tropomyosin 1 (alpha) |
| 7273 | TTN | titin |
| 7276 | TTR | transthyretin |
| 7414 | VCL | vinculin |
| 7840 | ALMS1 | Alstrom syndrome 1 |
| 8048 | CSRP3 | cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| 8557 | TCAP | titin-cap (telethonin) |
| 9499 | MYOT | myotilin |
| 9997 | SCO2 | SCO cytochrome oxidase deficient homolog 2 (yeast) |
| 10060 | ABCC9 | ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| 10102 | TSFM | Ts translation elongation factor, mitochondrial |
| 11155 | LDB3 | LIM domain binding 3 |
| 23417 | MLYCD | malonyl-CoA decarboxylase |
| 23459 | CMD1H | cardiomyopathy, dilated 1H (autosomal dominant) |
| 26580 | BSCL2 | Bernardinelli-Seip congenital lipodystrophy 2 (seipin) |
| 29078 | C6orf66 | chromosome 6 open reading frame 66 |
| 51422 | PRKAG2 | protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| 56945 | MRPS22 | mitochondrial ribosomal protein S22 |
| 65014 | CMD1K | cardiomyopathy, dilated 1K (autosomal dominant) |
| 79087 | ALG12 | asparagine-linked glycosylation 12, alpha-1,6-mannosyltransferase homolog (S. cerevisiae) |
| 79158 | GNPTAB | N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits |
| 85366 | MYLK2 | myosin light chain kinase 2 |
| 131118 | DNAJC19 | DnaJ (Hsp40) homolog, subfamily C, member 19 |
| 148738 | HFE2 | hemochromatosis type 2 (juvenile) |
| 197131 | UBR1 | ubiquitin protein ligase E3 component n-recognin 1 |
| 619468 | RCM2 | cardiomyopathy, familial restrictive, 2 |
| 664728 | CMD1Q | cardiomyopathy, dilated 1Q (autosomal dominant) |
Cardiomyopathy Test Set genes
MYBPC3 candidate gene (MYBPC3 + 99 nearest neighboring genes)
| Entrez Gene ID | Gene Symbol | Gene Name |
| 84680 | ACCS | 1-aminocyclopropane-1-carboxylate synthase homolog (Arabidopsis)(non-functional) |
| 390110 | ACCSL | 1-aminocyclopropane-1-carboxylate synthase homolog (Arabidopsis)(non-functional)-like |
| 53 | ACP2 | acid phosphatase 2, lysosomal |
| 79841 | AGBL2 | ATP/GTP binding protein-like 2 |
| 221120 | ALKBH3 | alkB, alkylation repair homolog 3 (E. coli) |
| 60529 | ALX4 | ALX homeobox 4 |
| 55626 | AMBRA1 | autophagy/beclin-1 regulator 1 |
| 84364 | ARFGAP2 | ADP-ribosylation factor GTPase activating protein 2 |
| 392 | ARHGAP1 | Rho GTPase activating protein 1 |
| 79096 | C11orf49 | chromosome 11 open reading frame 49 |
| 114900 | C1QTNF4 | C1q and tumor necrosis factor related protein 4 |
| 3732 | CD82 | CD82 molecule |
| 1132 | CHRM4 | cholinergic receptor, muscarinic 4 |
| 8534 | CHST1 | carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 |
| 9793 | CKAP5 | cytoskeleton associated protein 5 |
| 90993 | CREB3L1 | cAMP responsive element binding protein 3-like 1 |
| 1408 | CRY2 | cryptochrome 2 (photolyase-like) |
| 10658 | CUGBP1 | CUG triplet repeat, RNA binding protein 1 |
| 1643 | DDB2 | damage-specific DNA binding protein 2, 48kDa |
| 8525 | DGKZ | diacylglycerol kinase, zeta 104kDa |
| 2132 | EXT2 | exostoses (multiple) 2 |
| 2147 | F2 | coagulation factor II (thrombin) |
| 23360 | FNBP4 | formin binding protein 4 |
| 2346 | FOLH1 | folate hydrolase (prostate-specific membrane antigen) 1 |
| 120071 | GYLTL1B | glycosyltransferase-like 1B |
| 283254 | HARBI1 | harbinger transposase derived 1 |
| 55709 | KBTBD4 | kelch repeat and BTB (POZ) domain containing 4 |
| 9776 | KIAA0652 | KIAA0652 |
| 143678 | LOC143678 | hypothetical LOC143678 |
| 441601 | LOC441601 | septin 7 pseudogene |
| 646813 | LOC646813 | hypothetical LOC646813 |
| 4038 | LRP4 | low density lipoprotein receptor-related protein 4 |
| 8567 | MADD | MAP-kinase activating death domain |
| 9479 | MAPK8IP1 | mitogen-activated protein kinase 8 interacting protein 1 |
| 4192 | MDK | midkine (neurite growth-promoting factor 2) |
| 23788 | MTCH2 | mitochondrial carrier homolog 2 (C. elegans) |
| 4607 | MYBPC3 | myosin binding protein C, cardiac |
| 4722 | NDUFS3 | NADH dehydrogenase (ubiquinone) Fe-S protein 3, 30kDa (NADH-coenzyme Q reductase) |
| 10062 | NR1H3 | nuclear receptor subfamily 1, group H, member 3 |
| 23279 | NUP160 | nucleoporin 160kDa |
| 282770 | OR10AG1 | olfactory receptor, family 10, subfamily AG, member 1 |
| 81328 | OR4A15 | olfactory receptor, family 4, subfamily A, member 15 |
| 81327 | OR4A16 | olfactory receptor, family 4, subfamily A, member 16 |
| 403253 | OR4A47 | olfactory receptor, family 4, subfamily A, member 47 |
| 81318 | OR4A5 | olfactory receptor, family 4, subfamily A, member 5 |
| 119765 | OR4B1 | olfactory receptor, family 4, subfamily B, member 1 |
| 219429 | OR4C11 | olfactory receptor, family 4, subfamily C, member 11 |
| 283093 | OR4C12 | olfactory receptor, family 4, subfamily C, member 12 |
| 283092 | OR4C13 | olfactory receptor, family 4, subfamily C, member 13 |
| 81309 | OR4C15 | olfactory receptor, family 4, subfamily C, member 15 |
| 219428 | OR4C16 | olfactory receptor, family 4, subfamily C, member 16 |
| 256144 | OR4C3 | olfactory receptor, family 4, subfamily C, member 3 |
| 403257 | OR4C45 | olfactory receptor, family 4, subfamily C, member 45 |
| 119749 | OR4C46 | olfactory receptor, family 4, subfamily C, member 46 |
| 219432 | OR4C6 | olfactory receptor, family 4, subfamily C, member 6 |
| 81300 | OR4P4 | olfactory receptor, family 4, subfamily P, member 4 |
| 256148 | OR4S1 | olfactory receptor, family 4, subfamily S, member 1 |
| 219431 | OR4S2 | olfactory receptor, family 4, subfamily S, member 2 |
| 390113 | OR4X1 | olfactory receptor, family 4, subfamily X, member 1 |
| 119764 | OR4X2 | olfactory receptor, family 4, subfamily X, member 2 |
| 219447 | OR5AS1 | olfactory receptor, family 5, subfamily AS, member 1 |
| 390142 | OR5D13 | olfactory receptor, family 5, subfamily D, member 13 |
| 219436 | OR5D14 | olfactory receptor, family 5, subfamily D, member 14 |
| 390144 | OR5D16 | olfactory receptor, family 5, subfamily D, member 16 |
| 219438 | OR5D18 | olfactory receptor, family 5, subfamily D, member 18 |
| 338674 | OR5F1 | olfactory receptor, family 5, subfamily F, member 1 |
| 10798 | OR5I1 | olfactory receptor, family 5, subfamily I, member 1 |
| 282775 | OR5J2 | olfactory receptor, family 5, subfamily J, member 2 |
| 219437 | OR5L1 | olfactory receptor, family 5, subfamily L, member 1 |
| 26338 | OR5L2 | olfactory receptor, family 5, subfamily L, member 2 |
| 390155 | OR5T1 | olfactory receptor, family 5, subfamily T, member 1 |
| 219464 | OR5T2 | olfactory receptor, family 5, subfamily T, member 2 |
| 390154 | OR5T3 | olfactory receptor, family 5, subfamily T, member 3 |
| 390148 | OR5W2 | olfactory receptor, family 5, subfamily W, member 2 |
| 219469 | OR8H1 | olfactory receptor, family 8, subfamily H, member 1 |
| 390151 | OR8H2 | olfactory receptor, family 8, subfamily H, member 2 |
| 390152 | OR8H3 | olfactory receptor, family 8, subfamily H, member 3 |
| 120586 | OR8I2 | olfactory receptor, family 8, subfamily I, member 2 |
| 81168 | OR8J3 | olfactory receptor, family 8, subfamily J, member 3 |
| 390157 | OR8K1 | olfactory receptor, family 8, subfamily K, member 1 |
| 219473 | OR8K3 | olfactory receptor, family 8, subfamily K, member 3 |
| 219453 | OR8K5 | olfactory receptor, family 8, subfamily K, member 5 |
| 29763 | PACSIN3 | protein kinase C and casein kinase substrate in neurons 3 |
| 9409 | PEX16 | peroxisomal biogenesis factor 16 |
| 51317 | PHF21A | PHD finger protein 21A |
| 56981 | PRDM11 | PR domain containing 11 |
| 5702 | PSMC3 | proteasome (prosome, macropain) 26S subunit, ATPase, 3 |
| 114971 | PTPMT1 | protein tyrosine phosphatase, mitochondrial 1 |
| 5795 | PTPRJ | protein tyrosine phosphatase, receptor type, J |
| 5913 | RAPSN | receptor-associated protein of the synapse |
| 55343 | SLC35C1 | solute carrier family 35, member C1 |
| 91252 | SLC39A13 | solute carrier family 39 (zinc transporter), member 13 |
| 692108 | SNORD67 | small nucleolar RNA, C/D box 67 |
| 6688 | SPI1 | spleen focus forming virus (SFFV) proviral integration oncogene spi1 |
| 84767 | SPRYD5 | SPRY domain containing 5 |
| 57586 | SYT13 | synaptotagmin XIII |
| 9537 | TP53I11 | tumor protein p53 inducible protein 11 |
| 79097 | TRIM48 | tripartite motif-containing 48 |
| 90139 | TSPAN18 | tetraspanin 18 |
| 79797 | ZNF408 | zinc finger protein 408 |
67 Known celiac disease associated genes (Training set; compiled from OMIM and NCBI Entrez Gene)
| Entrez Gene ID | Gene Symbol | Gene Name |
| 100188846 | CELIAC6 | Celiac disease, susceptibility to, 6 |
| 100188870 | CELIAC8 | Celiac disease, susceptibility to, 8 |
| 100188871 | CELIAC9 | Celiac disease, susceptibility to, 9 |
| 100188872 | CELIAC10 | Celiac disease, susceptibility to, 10 |
| 100188873 | CELIAC11 | Celiac disease, susceptibility to, 11 |
| 100188874 | CELIAC12 | Celiac disease, susceptibility to, 12 |
| 100188875 | CELIAC13 | Celiac disease, susceptibility to, 13 |
| 10019 | SH2B3 | SH2B adaptor protein 3 |
| 100233228 | IDDM20 | Diabetes mellitus, insulin-dependent, 20 |
| 100233229 | IDDM21 | Diabetes mellitus, insulin-dependent, 21 |
| 117289 | TAGAP | T-cell activation RhoGTPase activating protein |
| 1234 | CCR5 | chemokine (C-C motif) receptor 5 |
| 132612 | ADAD1 | adenosine deaminase domain containing 1 (testis-specific) |
| 1493 | CTLA4 | cytotoxic T-lymphocyte-associated protein 4 |
| 2006 | ELN | elastin |
| 2022 | ENG | endoglin |
| 22914 | KLRK1 | killer cell lectin-like receptor subfamily K, member 1 |
| 2591 | GALNT3 | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3) |
| 26191 | PTPN22 | protein tyrosine phosphatase, non-receptor type 22 (lymphoid) |
| 2677 | GGCX | gamma-glutamyl carboxylase |
| 2717 | GLA | galactosidase, alpha |
| 2778 | GNAS | GNAS complex locus |
| 2969 | GTF2I | general transcription factor II, i |
| 29970 | SCHIP1 | schwannomin interacting protein 1 |
| 3117 | HLA-DQA1 | major histocompatibility complex, class II, DQ alpha 1 |
| 3119 | HLA-DQB1 | major histocompatibility complex, class II, DQ beta 1 |
| 317782 | CELIAC2 | celiac disease 2 |
| 33 | ACADL | acyl-Coenzyme A dehydrogenase, long chain |
| 338332 | CELIAC5 | Celiac disease, susceptibility to, 5 |
| 3558 | IL2 | interleukin 2 |
| 3592 | IL12A | interleukin 12A (natural killer cell stimulatory factor 1, cytotoxic lymphocyte maturation factor 1, p35) |
| 3600 | IL15 | interleukin 15 |
| 3710 | ITPR3 | inositol 1,4,5-triphosphate receptor, type 3 |
| 3826 | KMS | Kabuki mental retardation syndrome |
| 389015 | SLC9A4 | solute carrier family 9 (sodium/hydrogen exchanger), member 4 |
| 4026 | LPP | LIM domain containing preferred translocation partner in lipoma |
| 4276 | MICA | MHC class I polypeptide-related sequence A |
| 4436 | MSH2 | mutS homolog 2, colon cancer, nonpolyposis type 1 (E. coli) |
| 4547 | MTTP | microsomal triglyceride transfer protein |
| 4650 | MYO9B | myosin IXB |
| 4763 | NF1 | neurofibromin 1 |
| 4938 | OAS1 | 2',5'-oligoadenylate synthetase 1, 40/46kDa |
| 50854 | C6orf48 | chromosome 6 open reading frame 48 |
| 50943 | FOXP3 | forkhead box P3 |
| 5284 | PIGR | polymeric immunoglobulin receptor |
| 5621 | PRNP | prion protein |
| 5651 | PRSS7 | protease, serine, 7 (enterokinase) |
| 59067 | IL21 | interleukin 21 |
| 5996 | RGS1 | regulator of G-protein signaling 1 |
| 6023 | RMRP | RNA component of mitochondrial RNA processing endoribonuclease |
| 60498 | IGAN | IgA nephropathy |
| 6365 | SCZD3 | schizophrenia disorder 3 |
| 6401 | SELE | selectin E |
| 6402 | SELL | selectin L |
| 6833 | ABCC8 | ATP-binding cassette, sub-family C (CFTR/MRP), member 8 |
| 6890 | TAP1 | transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| 6927 | HNF1A | HNF1 homeobox A |
| 7052 | TGM2 | transglutaminase 2 (C polypeptide, protein-glutamine-gamma-glutamyltransferase) |
| 7053 | TGM3 | transglutaminase 3 (E polypeptide, protein-glutamine-gamma-glutamyltransferase) |
| 791087 | HHT4 | Telangiectasia, hereditary hemorrhagic, type 4 |
| 8074 | FGF23 | fibroblast growth factor 23 |
| 84162 | KIAA1109 | KIAA1109 |
| 8807 | IL18RAP | interleukin 18 receptor accessory protein |
| 8809 | IL18R1 | interleukin 18 receptor 1 |
| 9173 | IL1RL1 | interleukin 1 receptor-like 1 |
| 9365 | KL | klotho |
| 9569 | GTF2IRD1 | GTF2I repeat domain containing 1 |
Celiac disease Test Set genes for the following six candidate genes is compiled in a similar way as described for others previously.
SH2B3 candidate gene (SH2B3 + 99 nearest neighboring genes)
CCR3 candidate gene (CCR3 + 99 nearest neighboring genes)
IL18R1 candidate gene (IL18R1 + 99 nearest neighboring genes)
RGS1 candidate gene (RGS1 + 99 nearest neighboring genes)
TAGAP candidate gene (TAGAP + 99 nearest neighboring genes)
IL12A candidate gene (IL12A + 99 nearest neighboring genes)
12 Known Crohn's disease associated genes (Training set; compiled from OMIM and NCBI Entrez Gene)
| Entrez Gene ID | Gene Symbol | Gene Name |
| 183 | AGT | angiotensinogen (serpin peptidase inhibitor, clade A, member 8) |
| 1673 | DEFB4 | defensin, beta 4 |
| 3569 | IL6 | interleukin 6 (interferon, beta 2) |
| 3663 | IRF5 | interferon regulatory factor 5 |
| 5243 | ABCB1 | ATP-binding cassette, sub-family B (MDR/TAP), member 1 |
| 6583 | SLC22A4 | solute carrier family 22 (organic cation/ergothioneine transporter), member 4 |
| 7466 | WFS1 | Wolfram syndrome 1 (wolframin) |
| 9231 | DLG5 | discs, large homolog 5 (Drosophila) |
| 55054 | ATG16L1 | ATG16 autophagy related 16-like 1 (S. cerevisiae) |
| 64127 | NOD2 | nucleotide-binding oligomerization domain containing 2 |
| 149233 | IL23R | interleukin 23 receptor |
| 256815 | C10orf67 | chromosome 10 open reading frame 67 |
Crohn's disease Test Set genes for the following seven candidate genes is compiled in a similar way as described for others previously.
MST1 candidate gene (MST1 + 99 nearest neighboring genes)
NKX2-3 candidate gene (NKX2-3 + 99 nearest neighboring genes)
IRGM candidate gene (IRGM + 99 nearest neighboring genes)
NLRP3 candidate gene (NLRP3 + 99 nearest neighboring genes)
IL12B candidate gene (IL12B + 99 nearest neighboring genes)
STAT3 candidate gene (STAT3 + 99 nearest neighboring genes)
PTPN2 candidate gene (PTPN2 + 99 nearest neighboring genes)
63 Known obesity associated genes (Training set; compiled from OMIM and NCBI Entrez Gene)
| Entrez Gene ID | Gene Symbol | Gene Name |
| 154 | ADRB2 | adrenergic, beta-2-, receptor, surface |
| 155 | ADRB3 | adrenergic, beta-3-, receptor |
| 181 | AGRP | agouti related protein homolog (mouse) |
| 338 | APOB | apolipoprotein B (including Ag(x) antigen) |
| 627 | BDNF | brain-derived neurotrophic factor |
| 1270 | CNTF | ciliary neurotrophic factor |
| 1636 | ACE | angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 |
| 1646 | AKR1C2 | aldo-keto reductase family 1, member C2 (dihydrodiol dehydrogenase 2; bile acid binding protein; 3-alpha hydroxysteroid dehydrogenase, type III) |
| 1815 | DRD4 | dopamine receptor D4 |
| 2692 | GHRHR | growth hormone releasing hormone receptor |
| 2784 | GNB3 | guanine nucleotide binding protein (G protein), beta polypeptide 3 |
| 2908 | NR3C1 | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| 3290 | HSD11B1 | hydroxysteroid (11-beta) dehydrogenase 1 |
| 3557 | IL1RN | interleukin 1 receptor antagonist |
| 3569 | IL6 | interleukin 6 (interferon, beta 2) |
| 3643 | INSR | insulin receptor |
| 3952 | LEP | leptin |
| 3953 | LEPR | leptin receptor |
| 3972 | LHB | luteinizing hormone beta polypeptide |
| 4023 | LPL | lipoprotein lipase |
| 4159 | MC3R | melanocortin 3 receptor |
| 4663 | NA | neurocanthocytosis |
| 4852 | NPY | neuropeptide Y |
| 4915 | NTRK2 | neurotrophic tyrosine kinase, receptor, type 2 |
| 5122 | PCSK1 | proprotein convertase subtilisin/kexin type 1 |
| 5167 | ENPP1 | ectonucleotide pyrophosphatase/phosphodiesterase 1 |
| 5443 | POMC | proopiomelanocortin |
| 5465 | PPARA | peroxisome proliferator-activated receptor alpha |
| 5468 | PPARG | peroxisome proliferator-activated receptor gamma |
| 5506 | PPP1R3A | protein phosphatase 1, regulatory (inhibitor) subunit 3A |
| 6197 | RPS6KA3 | ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| 6303 | SAT1 | spermidine/spermine N1-acetyltransferase 1 |
| 6492 | SIM1 | single-minded homolog 1 (Drosophila) |
| 6927 | HNF1A | HNF1 homeobox A |
| 7068 | THRB | thyroid hormone receptor, beta (erythroblastic leukemia viral (v-erb-a) oncogene homolog 2, avian) |
| 7350 | UCP1 | uncoupling protein 1 (mitochondrial, proton carrier) |
| 7351 | UCP2 | uncoupling protein 2 (mitochondrial, proton carrier) |
| 7352 | UCP3 | uncoupling protein 3 (mitochondrial, proton carrier) |
| 7492 | WTS | Wilson-Turner X-linked mental retardation syndrome |
| 8195 | MKKS | McKusick-Kaufman syndrome |
| 8203 | OQTL | Obesity quantitative trait locus |
| 8422 | MEHMO | mental retardation, epileptic seizures, hypogonadism and -genitalism, microcephaly and obesity syndrome |
| 8431 | NR0B2 | nuclear receptor subfamily 0, group B, member 2 |
| 8660 | IRS2 | insulin receptor substrate 2 |
| 9563 | H6PD | hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| 9607 | CARTPT | CART prepropeptide |
| 9672 | SDC3 | syndecan 3 |
| 11132 | CAPN10 | calpain 10 |
| 11254 | SLC6A14 | solute carrier family 6 (amino acid transporter), member 14 |
| 23590 | PDSS1 | prenyl (decaprenyl) diphosphate synthase, subunit 1 |
| 50940 | PDE11A | phosphodiesterase 11A |
| 51738 | GHRL | ghrelin/obestatin prepropeptide |
| 54903 | MKS1 | Meckel syndrome, type 1 |
| 56694 | OB10P | obesity, susceptibility to, on chromosome 10p |
| 65076 | AOMS1 | Abdominal obesity-metabolic syndrome QTL1 |
| 65077 | AOMS2 | abdominal obesity-metabolic syndrome QTL2 |
| 80184 | CEP290 | centrosomal protein 290kDa |
| 116519 | APOA5 | apolipoprotein A-V |
| 133522 | PPARGC1B | peroxisome proliferator-activated receptor gamma, coactivator 1 beta |
| 255239 | ANKK1 | ankyrin repeat and kinase domain containing 1 |
| 353126 | OB10Q | Obesity, susceptibility to, on chromosome 10q |
| 404683 | OB4 | Obesity, susceptibility to, on chromosome 4 |
| 79068 | FTO | fat mass and obesity associated |
Obesity Test Set genes
MC4R candidate gene (MC4R + 99 nearest neighboring genes)
| Entrez Gene ID | Gene Symbol | Gene Name |
| 10449 | ACAA2 | acetyl-Coenzyme A acyltransferase 2 |
| 115701 | ALPK2 | alpha-kinase 2 |
| 5205 | ATP8B1 | ATPase, class I, type 8B, member 1 |
| 374868 | ATP9B | ATPase, class II, type 9B |
| 596 | BCL2 | B-cell CLL/lymphoma 2 |
| 220134 | C18orf24 | chromosome 18 open reading frame 24 |
| 284254 | C18orf26 | chromosome 18 open reading frame 26 |
| 125704 | C18orf51 | chromosome 18 open reading frame 51 |
| 162681 | C18orf54 | chromosome 18 open reading frame 54 |
| 29090 | C18orf55 | chromosome 18 open reading frame 55 |
| 284274 | C18orf62 | chromosome 18 open reading frame 62 |
| 147381 | CBLN2 | cerebellin 2 precursor |
| 147372 | CCBE1 | collagen and calcium binding EGF domains 1 |
| 79839 | CCDC102B | coiled-coil domain containing 102B |
| 220136 | CCDC11 | coiled-coil domain containing 11 |
| 80323 | CCDC68 | coiled-coil domain containing 68 |
| 10666 | CD226 | CD226 molecule |
| 28513 | CDH19 | cadherin 19, type 2 |
| 28316 | CDH20 | cadherin 20, type 2 |
| 1005 | CDH7 | cadherin 7, type 2 |
| 84735 | CNDP1 | carnosine dipeptidase 1 (metallopeptidase M20 family) |
| 55748 | CNDP2 | CNDP dipeptidase 2 (metallopeptidase M20 family) |
| 339302 | CPLX4 | complexin 4 |
| 9150 | CTDP1 | CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 |
| 30827 | CXXC1 | CXXC finger 1 (PHD domain) |
| 1528 | CYB5A | cytochrome b5 type A (microsomal) |
| 1630 | DCC | deleted in colorectal carcinoma |
| 220164 | DOK6 | docking protein 6 |
| 92126 | DSEL | dermatan sulfate epimerase-like |
| 55520 | ELAC1 | elaC homolog 1 (E. coli) |
| 201456 | FBXO15 | F-box protein 15 |
| 2235 | FECH | ferrochelatase (protoporphyria) |
| 2587 | GALR1 | galanin receptor 1 |
| 2922 | GRP | gastrin-releasing peptide |
| 284293 | HMSD | histocompatibility (minor) serpin domain containing |
| 2531 | KDSR | 3-ketodihydrosphingosine reductase |
| 57614 | KIAA1468 | KIAA1468 |
| 9388 | LIPG | lipase, endothelial |
| 3998 | LMAN1 | lectin, mannose-binding, 1 |
| 284276 | LOC284276 | hypothetical LOC284276 |
| 400657 | LOC400657 | hypothetical LOC400657 |
| 10892 | MALT1 | mucosa associated lymphoid tissue lymphoma translocation gene 1 |
| 5596 | MAPK4 | mitogen-activated protein kinase 4 |
| 4152 | MBD1 | methyl-CpG binding domain protein 1 |
| 8932 | MBD2 | methyl-CpG binding domain protein 2 |
| 4155 | MBP | myelin basic protein |
| 4160 | MC4R | melanocortin 4 receptor |
| 4200 | ME2 | malic enzyme 2, NAD(+)-dependent, mitochondrial |
| 51320 | MEX3C | mex-3 homolog C (C. elegans) |
| 83876 | MRO | maestro |
| 4645 | MYO5B | myosin VB |
| 4677 | NARS | asparaginyl-tRNA synthetase |
| 23327 | NEDD4L | neural precursor cell expressed, developmentally down-regulated 4-like |
| 81832 | NETO1 | neuropilin (NRP) and tolloid (TLL)-like 1 |
| 4772 | NFATC1 | nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 |
| 9480 | ONECUT2 | one cut homeobox 2 |
| 23239 | PHLPP | PH domain and leucine rich repeat protein phosphatase |
| 23556 | PIGN | phosphatidylinositol glycan anchor biosynthesis, class N |
| 5366 | PMAIP1 | phorbol-12-myristate-13-acetate-induced protein 1 |
| 11201 | POLI | polymerase (DNA directed) iota |
| 5874 | RAB27B | RAB27B, member RAS oncogene family |
| 30062 | RAX | retina and anterior neural fold homeobox |
| 220441 | RNF152 | ring finger protein 152 |
| 25914 | RTTN | rotatin |
| 27164 | SALL3 | sal-like 3 (Drosophila) |
| 677769 | SCARNA17 | small Cajal body-specific RNA 17 |
| 90701 | SEC11C | SEC11 homolog C (S. cerevisiae) |
| 5273 | SERPINB10 | serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
| 89778 | SERPINB11 | serpin peptidase inhibitor, clade B (ovalbumin), member 11 |
| 89777 | SERPINB12 | serpin peptidase inhibitor, clade B (ovalbumin), member 12 |
| 5275 | SERPINB13 | serpin peptidase inhibitor, clade B (ovalbumin), member 13 |
| 5055 | SERPINB2 | serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
| 6317 | SERPINB3 | serpin peptidase inhibitor, clade B (ovalbumin), member 3 |
| 6318 | SERPINB4 | serpin peptidase inhibitor, clade B (ovalbumin), member 4 |
| 5268 | SERPINB5 | serpin peptidase inhibitor, clade B (ovalbumin), member 5 |
| 8710 | SERPINB7 | serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| 5271 | SERPINB8 | serpin peptidase inhibitor, clade B (ovalbumin), member 8 |
| 4089 | SMAD4 | SMAD family member 4 |
| 677819 | SNORA37 | small nucleolar RNA, H/ACA box 37 |
| 26791 | SNORD58A | small nucleolar RNA, C/D box 58A |
| 26790 | SNORD58B | small nucleolar RNA, C/D box 58B |
| 100124516 | SNORD58C | small nucleolar RNA, C/D box 58C |
| 9306 | SOCS6 | suppressor of cytokine signaling 6 |
| 51046 | ST8SIA3 | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| 147323 | STARD6 | StAR-related lipid transfer (START) domain containing 6 |
| 6925 | TCF4 | transcription factor 4 |
| 8792 | TNFRSF11A | tumor necrosis factor receptor superfamily, member 11a, NFKB activator |
| 10194 | TSHZ1 | teashirt zinc finger homeobox 1 |
| 54495 | TXNDC10 | thioredoxin domain containing 10 |
| 9352 | TXNL1 | thioredoxin-like 1 |
| 619491 | U58 | U58 small nucleolar RNA |
| 9525 | VPS4B | vacuolar protein sorting 4 homolog B (S. cerevisiae) |
| 23335 | WDR7 | WD repeat domain 7 |
| 284273 | ZADH2 | zinc binding alcohol dehydrogenase domain containing 2 |
| 54877 | ZCCHC2 | zinc finger, CCHC domain containing 2 |
| 7776 | ZNF236 | zinc finger protein 236 |
| 55628 | ZNF407 | zinc finger protein 407 |
| 9658 | ZNF516 | zinc finger protein 516 |
| 55205 | ZNF532 | zinc finger protein 532 |
| 642250 | hCG_39912 | hCG39912 |
1. Le-Niculescu, H., Patel, S.D., Bhat, M., Kuczenski, R.,
Faraone, S.V., Tsuang, M.T., McMahon, F.J., Schork, N.J.,
Nurnberger, J.I., Jr. and Niculescu, A.B., 3rd. (2008)
Convergent functional genomics of genome-wide association data
for bipolar disorder: Comprehensive identification of candidate
genes, pathways and mechanisms. Am J Med Genet B Neuropsychiatr
Genet.
2. Dhandapany, P.S., Sadayappan, S., Xue, Y., Powell, G.T.,
Rani, D.S., Nallari, P., Rai, T.S., Khullar, M., Soares, P.,
Bahl, A. et al. (2009) A common MYBPC3 (cardiac myosin binding
protein C) variant associated with cardiomyopathies in South
Asia. Nature genetics.
3. Hunt, K.A., Zhernakova, A., Turner, G., Heap, G.A., Franke,
L., Bruinenberg, M., Romanos, J., Dinesen, L.C., Ryan, A.W.,
Panesar, D. et al. (2008) Newly identified genetic risk
variants for celiac disease related to the immune response.
Nature genetics, 40, 395-402.
4. Fisher, S.A., Tremelling, M., Anderson, C.A., Gwilliam, R.,
Bumpstead, S., Prescott, N.J., Nimmo, E.R., Massey, D.,
Berzuini, C., Johnson, C. et al. (2008) Genetic determinants of
ulcerative colitis include the ECM1 locus and five loci
implicated in Crohn's disease. Nature genetics, 40,
710-712.
5. Villani, A.C., Lemire, M., Fortin, G., Louis, E.,
Silverberg, M.S., Collette, C., Baba, N., Libioulle, C.,
Belaiche, J., Bitton, A. et al. (2009) Common variants in the
NLRP3 region contribute to Crohn's disease susceptibility.
Nature genetics, 41, 71-76. Barrett, J.C., Hansoul, S.,
Nicolae, D.L., Cho, J.H., Duerr, R.H., Rioux, J.D., Brant,
S.R., Silverberg, M.S., Taylor, K.D., Barmada, M.M. et al.
(2008) Genome-wide association defines more than 30 distinct
susceptibility loci for Crohn's disease. Nature genetics, 40,
955-962.
6. Franke, A., Balschun, T., Karlsen, T.H., Hedderich, J., May,
S., Lu, T., Schuldt, D., Nikolaus, S., Rosenstiel, P.,
Krawczak, M. et al. (2008) Replication of signals from recent
studies of Crohn's disease identifies previously unknown
disease loci for ulcerative colitis. Nature genetics, 40,
713-715.
7. Renstrom, F., Payne, F., Nordstrom, A., Brito, E.C.,
Rolandsson, O., Hallmans, G., Barroso, I., Nordstrom, P. and
Franks, P.W. (2009) Replication and extension of genome-wide
association study results for obesity in 4,923 adults from
Northern Sweden. Hum Mol Genet.